Ogataea polymorpha
| Ogataea polymorpha | |
|---|---|
| Scientific classification | |
| Kingdom: | |
| Division: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | O. polymorpha
|
| Binomial name | |
| Ogataea polymorpha (Morais & M.H.Maia) Y.Yamada, K.Maeda & Mikata
| |
| Synonyms [1] | |
| |
Ogataea polymorpha is a methylotrophic yeast with unusual characteristics. It is used as a protein factory for pharmaceuticals.
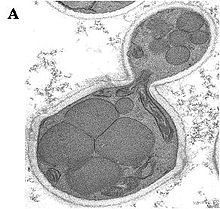

Ogataea polymorpha belongs to a limited number of methylotrophic yeast species – yeasts that can grow on methanol. The range of methylotrophic yeasts includes Candida boidinii, Pichia methanolica, Pichia pastoris and Ogataea polymorpha. O. polymorpha is taxonomically a species of the family Saccharomycetaceae.
Strains
[edit]Three O. polymorpha strains, identified in the 1950s, are known. They have unclear relationships and are of independent origins. They are found in soil samples, the gut of insects or in spoiled concentrated orange juice. They exhibit different features and are used in basic research and to recombinant protein production:[2]
- strain CBS4732 (CCY38-22-2; ATCC34438, NRRL-Y-5445)
- strain DL-1 (NRRL-Y-7560; ATCC26012)
- strain NCYC495 (CBS1976; ATAA14754, NRLL-Y-1798)
Strains CBS4732 and NCYY495 can be mated whereas strain DL-1 cannot be mated with the other two. Strains CBS4732 and DL-1 are employed for recombinant protein production, strain NCYC495 is mainly used for the study of nitrate assimilation. The entire genome of strain CBS4732 has completely been sequenced.[citation needed]
Ogataea polymorpha is a thermo-tolerant microorganism with some strains growing at temperatures above 50 °C (122 °F). The organism is able to assimilate nitrate and can grow on a range of carbon sources in addition to methanol. Cells grown under conditions of elevated temperature accumulate a sugar named trehalose (this sugar is usually found in insects) as thermo-protective compound. It was shown that trehalose synthesis is not required for growth under these conditions, but for acquisition of thermotolerance. The synthetic steps for trehalose synthesis have been detailed for O. polymorpha, and TPS1, the key enzyme gene of this pathway, has been isolated and characterized.[citation needed]
All methylotrophic yeasts share an identical methanol utilization pathway (Fig. 1). Growth on methanol is accompanied by a massive proliferation of cell organelles named peroxisomes in which the initial enzymatic steps of this pathway take place. O. polymorpha is model organism to study all aspects of peroxisomal functions and the underlying molecular biology. During growth on methanol key enzymes of the methanol metabolism are present in high amounts. An especially high abundance can be observed for enzymes called MOX (methanol oxidase), FMDH (formate dehydrogenase), and DHAS (dihydroxyacetone synthase). Their presence is regulated at the transcriptional level of the respective genes. In the related species C. boidinii, P. methanolica, and P. pastoris this gene expression strictly depends on the presence of methanol or methanol derivatives, whereas in O. polymorpha strong expression is elicited by appropriate levels of glycerol or under conditions of glucose starvation. O. polymorpha produces glycoproteins with two types of sugar chains, N- and O-linked glycans are attached to protein. Studies on the structure of N-linked chains have revealed a certain average length (Man8-12GlcNAc2) with terminal alpha-1,2-linked mannose residues, and not with allergenic terminal alpha-1,3-linked mannose residues as found in other yeasts, especially in the baker’s yeast Saccharomyces cerevisiae.[citation needed]
Biotechnological applications
[edit]Ogataea polymorpha with its unusual characteristics provides an excellent platform for the gene technological production of proteins, especially of pharmaceuticals like insulin for treatment of diabetes, hepatitis B vaccines or IFNalpha-2a for the treatment of hepatitis C. Derivatives of both CBS4732 and DL-1 are employed in the production of such recombinant compounds. Further yeasts employed for this application are Pichia pastoris, Arxula adeninivorans and Saccharomyces cerevisiae and others.
Like other yeasts, O. polymorpha is a microorganism that can be cultured in large fermenters to high cell densities within a short time. It is a safe organism in not containing pyrogens, pathogens or viral inclusions. It can release compounds into a culture medium as it has all the components required for secretion (this is for instance not the case with bacteria like Escherichia coli). It can provide attractive genetic components for an efficient production of proteins.
In Fig. 2 the general design of a vector (a genetic vehicle to transform a yeast strain into a genetically engineered protein producer). It must contain several genetic elements: 1. A selection marker, required to select a transformed strain from an untransformed background –this can be done if for instance such an element enables a deficient strain to grow under culturing conditions void of a certain compound like a particular amino acid that cannot be produced by the deficient strain). 2. Certain elements to propagate and to target the foreign DNA to the chromosome of the yeast (ARS and/or rDNA sequence). 3. A segment responsible for the production of the desired protein compound a so-called expression cassette. Such a cassette is made up by a sequence of regulatory elements, a promoter that controls, how much and under which circumstances a following gene sequence is transcribed and as a consequence how much protein is eventually made. This means that the segment following the promoter is variable depending on the desired product – it could be a sequence determining the amino acids for insulin, for hepatitis B vaccine or for interferon. The expression cassette is terminated by a following terminator sequence that provides a proper stop of the transcription. The promoter elements of the O. polymorpha system are derived from genes that are highly expressed, from instance from the MOX gene, the FMD gene or the TPS1 gene mentioned before. They are not only very strong, but can also be regulated by certain addition of carbon sources like sugar, methanol or glycerol.

In 2000 an informal society of scientists was founded named HPWN (Hansenula polymorpha worldwide network) founded by Marten Veenhuis, Groningen, and Gerd Gellissen, Düsseldorf. Every two years meetings are held.
The attractiveness of the O. polymorpha platform is commercially exploited by several biotech companies for the development of production processes, among others by PharmedArtis, located in Aachen, Germany and the Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK).
References
[edit]- ^ "Ogataea polymorpha (Morais & M.H. Maia) Y. Yamada, K. Maeda & Mikata, Biosc., Biotechn., Biochem. 58(7): 1254 (1994)". Index Fungorum. Retrieved June 27, 2011.
- ^ Chang, Jia; Bei, Jinlong; Shao, Qi; Wang, Hemu; Fan, Huan; Yau, Tung On; Bu, Wenjun; Ruan, Jishou; Wei, Dongsheng; Gao, Shan (4 April 2022). "Full-Length Genome of an Ogataea polymorpha Strain CBS4732 ura3Δ Reveals Large Duplicated Segments in Subtelomeric Regions". Frontiers in Microbiology. 13: 855666. doi:10.3389/fmicb.2022.855666. PMC 9019687. PMID 35464988.
Bibliography
[edit]- Massoud Ramezani-Rad; Cornelis P. Hollenberg; Juergen Lauber; Holger Wedler; Eike Griess; Christian Wagner; Kaj Albermann; Jean Hani; Michael Piontek; Ulrike Dahlems; Gerd Gellissen (2003). "The Hansenula polymorpha (strain CBS4732) genome - sequencing and analysis". FEMS Yeast Research. 4 (2): 207–215. doi:10.1016/S1567-1356(03)00125-9. PMID 14613885.
- Gerd Gellissen; Gotthard Kunze; Claude Gaillardin; James M. Cregg; Enrico Berardi; Marten Veenhuis; Ida van der Klei (2005). "New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and dimorphic Arxula adeninivorans and Yarrowia lipolytica – a comparison". FEMS Yeast Research. 5 (11): 1079–1096. doi:10.1016/j.femsyr.2005.06.004. PMID 16144775.
- Gerd Gellissen, ed. (2002). Hansenula polymorpha – biology and applications. Weinheim: Wiley-VCH. ISBN 3-527-30341-3.
- Gerd Gellissen, ed. (2005). Production of recombinant proteins – novel microbial and eukaryotic expression systems. Weinheim: Wiley-VCH. ISBN 3-527-31036-3.
