Human genetic resistance to malaria: Difference between revisions
ClueBot NG (talk | contribs) m Reverting possible vandalism by Little.tone52 to version by 98.69.197.32. False positive? Report it. Thanks, ClueBot NG. (1735366) (Bot) |
Hempelmann (talk | contribs) →Sickle-cell: new refereence |
||
| Line 27: | Line 27: | ||
The modern phase of research on this disorder was initiated by the famous chemist [[Linus Pauling]] in 1949. Pauling postulated that the [[hemoglobin]] (Hb) in sickle-cell disease is abnormal; when deoxygenated it polymerizes into long, thin, helical rods that distort the red cell into a sickle shape. In his laboratory, electrophoretic studies showed that sickle-cell Hb (S) is indeed abnormal, having at physiological [[pH]] a lower negative charge than normal adult human Hb (A).<ref>{{cite journal | doi = 10.1126/science.110.2865.543 | author = Pauling L, Itano HA, Singer SJ, Wells IC | title = Sickle cell anemia, a molecular disease| journal = Science | volume = 110| issue = 2865 | pages = 543–548| year = 1949 |url = http://osulibrary.oregonstate.edu/specialcollections/coll/pauling/blood/papers/1949p.15.html |pmid = 15395398 | bibcode=1949Sci...110..543P}}</ref> In sickle-cell trait carriers there is a nearly equal amount of HbA and HbS, whereas in persons with sickle-cell disease nearly all the Hb is of the S type, apart from a small amount of fetal Hb. These observations showed that most patients with sickle-cell disease are homozygous for the gene encoding HbS, while trait carriers are heterozygous for this gene. Persons inheriting a sickle-cell gene and another mutant at the same locus, e.g. a thalassemia gene, can also have a variant form of sickle-cell disease. Pauling also introduced the term "molecular disease", which, together with ''[[molecular medicine]]'', has become widely used. |
The modern phase of research on this disorder was initiated by the famous chemist [[Linus Pauling]] in 1949. Pauling postulated that the [[hemoglobin]] (Hb) in sickle-cell disease is abnormal; when deoxygenated it polymerizes into long, thin, helical rods that distort the red cell into a sickle shape. In his laboratory, electrophoretic studies showed that sickle-cell Hb (S) is indeed abnormal, having at physiological [[pH]] a lower negative charge than normal adult human Hb (A).<ref>{{cite journal | doi = 10.1126/science.110.2865.543 | author = Pauling L, Itano HA, Singer SJ, Wells IC | title = Sickle cell anemia, a molecular disease| journal = Science | volume = 110| issue = 2865 | pages = 543–548| year = 1949 |url = http://osulibrary.oregonstate.edu/specialcollections/coll/pauling/blood/papers/1949p.15.html |pmid = 15395398 | bibcode=1949Sci...110..543P}}</ref> In sickle-cell trait carriers there is a nearly equal amount of HbA and HbS, whereas in persons with sickle-cell disease nearly all the Hb is of the S type, apart from a small amount of fetal Hb. These observations showed that most patients with sickle-cell disease are homozygous for the gene encoding HbS, while trait carriers are heterozygous for this gene. Persons inheriting a sickle-cell gene and another mutant at the same locus, e.g. a thalassemia gene, can also have a variant form of sickle-cell disease. Pauling also introduced the term "molecular disease", which, together with ''[[molecular medicine]]'', has become widely used. |
||
The next major advance was the discovery by [[Vernon Ingram]] in 1959 that HbS differs from HbA by only a single amino-acid substitution in the β-polypeptide chain (β6Glu → Val).<ref>{{cite journal | doi = 10.1016/0006-3002(59)90183-0 | author = Ingram VM. | title = Abnormal human haemoglobins. III. The chemical difference between normal and sickle cell haemoglobins| journal = Biochim Biophys Acta | volume = 36 | pages = 402–411| year = 1959 |pmid = 13852872 | issue = 2}}</ref> It was later established that this results from a substitution of [[thymine]] for [[adenine]] in the [[DNA]] [[codon]] (GAG → GTG). This was the first example in any species of the effects of a [[mutation]] on a [[protein]]. Sickle Hb induces the expression of heme oxygenase-1 in hematopoietic cells. [[Carbon monoxide]], a byproduct of heme catabolism by [[heme oxygenase]]-1, prevents an accumulation of circulating free heme after ''[[Plasmodium]]'' infection, suppressing the pathogenesis of experimental cerebral malaria.<ref>{{cite journal | author = Ferreira A, Marguti I, Bechmann I, Jeney V, Chora A, Palha NR, Rebelo S, Henri A, Beuzard Y, Soares MP| title = Sickle hemoglobin confers tolerance to Plasmodium infection| journal = Cell | volume = 145| issue = 3| pages = 398–409| year = 2011 |url = http://saturn.med.nyu.edu/research/mp/littmanlab/Immunology_Journal_Club/Papers_files/Ferreira.pdf|pmid = 21529713 | doi=10.1016/j.cell.2011.03.049}}</ref> |
The next major advance was the discovery by [[Vernon Ingram]] in 1959 that HbS differs from HbA by only a single amino-acid substitution in the β-polypeptide chain (β6Glu → Val).<ref>{{cite journal | doi = 10.1016/0006-3002(59)90183-0 | author = Ingram VM. | title = Abnormal human haemoglobins. III. The chemical difference between normal and sickle cell haemoglobins| journal = Biochim Biophys Acta | volume = 36 | pages = 402–411| year = 1959 |pmid = 13852872 | issue = 2}}</ref> It was later established that this results from a substitution of [[thymine]] for [[adenine]] in the [[DNA]] [[codon]] (GAG → GTG). This was the first example in any species of the effects of a [[mutation]] on a [[protein]]. Sickle Hb induces the expression of heme oxygenase-1 in hematopoietic cells. [[Carbon monoxide]], a byproduct of heme catabolism by [[heme oxygenase]]-1, prevents an accumulation of circulating free heme after ''[[Plasmodium]]'' infection, suppressing the pathogenesis of experimental cerebral malaria.<ref>{{cite journal | author = Ferreira A, Marguti I, Bechmann I, Jeney V, Chora A, Palha NR, Rebelo S, Henri A, Beuzard Y, Soares MP| title = Sickle hemoglobin confers tolerance to Plasmodium infection| journal = Cell | volume = 145| issue = 3| pages = 398–409| year = 2011 |url = http://saturn.med.nyu.edu/research/mp/littmanlab/Immunology_Journal_Club/Papers_files/Ferreira.pdf|pmid = 21529713 | doi=10.1016/j.cell.2011.03.049}}</ref> Other mechanisms, such as enhanced tolerance to disease mediated by HO-1 and reduced parasitic growth due to translocation of host micro-RNA into the parasite, have been described.<ref>{{cite journal | author = Gong L, Parikh S, Rosenthal PJ, Greenhouse B | title = Biochemical and immunological mechanisms by which sickle cell trait protects against malaria | journal = Malaria Journal | volume = 12| pages = 317| year = 2013 |url = http://www.malariajournal.com/content/pdf/1475-2875-12-317.pdf |pmid = 24025776}}</ref> |
||
==== Distribution of the sickle-cell gene ==== |
==== Distribution of the sickle-cell gene ==== |
||
Revision as of 18:48, 22 September 2013
Genetic resistance to malaria occurs through both modifications of the immune system that enhance immunity to this infection and also by changes in human red blood cells that hinder the malaria parasite's ability to invade and replicate within these cells. Host resistance to malaria therefore involves not only blood cell genes such as abnormal hemoglobins, Glucose-6-phosphate dehydrogenase deficiency, and Duffy antigens, which provide innate resistance, but also genes involved in immunity such as the major histocompatibility complex genes, which regulate adaptive immune responses.[1] The resistance provided by modified blood cells aids survival through the dangerous years of early childhood, while the potent protection mediated by adaptive immune responses is more important in older children and adults living where malaria is endemic.
Malaria has placed the strongest known selective pressure on the human genome since the origination of agriculture within the past 10,000 years.[2][3] Several inherited variants in erythrocytes have become common in formerly malarious parts of the world as a result of selection exerted by this parasite.[4] This selection was historically important as the first documented example of disease as an agent of natural selection in humans. It was also the first example of genetically controlled innate immunity that operates early in the course of infections, preceding adaptive immunity which exerts effects after several days. In malaria, as in other diseases, innate immunity leads into, and stimulates, adaptive immunity.
Population genetics
Evolution results from changes in gene frequencies in populations. The rates at which gene frequencies change, and the conditions under which they remain stable, were defined by the mathematical analyses of Ronald Fisher (The Genetical Theory of Natural Selection)[5] and John Burdon Sanderson Haldane in the United Kingdom and Sewall Wright in the U.S.A. in the decade preceding the Second World War. Individuals of different genotypes vary in "fitness", the probability that their genes will be passed on to the next generation. The term includes survival through reproductive age and relative fertility. Under some conditions genetic diversity (polymorphism) is stable, for example when a heterozygote has a fitness greater than that of either homozygote, whereas under other conditions polymorphism is unstable. Persons homozygous for abnormal hemoglobin (Hb) genes often have fitnesses lower than those with normal Hb, while heterozygotes have a greater fitness because of relative resistance to malaria, thereby maintaining stable polymorphisms in malarious environments.
Natural history of infections

In 2006 the World Health Organization estimated that there were about 250 million cases of malaria with 880,000 deaths. Approximately 90% of those who died were children in Africa infected with Plasmodium falciparum. Where this parasite is endemic young children have repeated malaria attacks. These are initially severe, and can be fatal, usually because of serious anemia or cerebral malaria. Repeated malaria infections strengthen adaptive immunity and broaden its effects against parasites expressing different surface antigens. By school age most children have developed efficacious adaptive immunity against malaria. These observations raise questions about mechanisms that favor the survival of most children in Africa while allowing some to develop potentially lethal infections. Evidence has accumulated that the first line of defense against malaria is provided by genetically controlled innate resistance, mainly exerted by abnormal hemoglobins and glucose-6-phosphate dehydrogenase deficiency. In malaria, as in other infections,[7] innate immune responses lead into, and stimulate, adaptive immune responses. However, the potent effect of genetically controlled innate resistance is reflected in the probability of survival of young children in malarious environments. It is necessary to study innate immunity in the susceptible age group, younger than four years; in older children and adults the effects of innate immunity are overshadowed by those of adaptive immunity. It is also necessary to study populations in which random use of antimalarial drugs does not occur.
Sickle-cell
The first report of sickle cell anaemia may have been in a report in 1846 where the autopsy report of an executed runaway slave was discussed.[8] The author noted the curious absence of a spleen in this case.
In 1910 a Chicago physician, James B. Herrick, reported the presence of sickle cells in the blood of an anaemic dental student, Walter Clement Noel.[9] These cells had first been observed by his intern Ernest Irons while they were treating Noel in 1904.
An association with pigmented gall stones was noted in 1911 by Washborn. A genetic basis for this disease was proposed in 1915 by Cook and Meyer. The disease was named sickle cell anaemia in 1922 by Verne Mason after several additional cases were reported. All the known cases had been reported in blacks and he concluded that this disease was confined to those of black African descent. The heterozygous condition was independently recognised in 1923 by Huck and Syndestrickler. Syndestrickler also was the first to note the splenic atrophy that occurs in this condition. It was recognised as a Mendelian autosomal characteristic by Taliaffero and Huck also in 1923.[10] A predisposition to pneumonia was noted in 1924 by Graham. The concept of progressive splenic atrophy was proposed by Hahn and Gilespie in 1927. Pneumococcal meningitis in this condition was first reported in 1928 by Wollstein and Kriedel but it was not until 1966 that the association between splenic atrophy and infection was made by Robinson and Watson.
In 1927 Vernon Hahn and Elizabeth Biermann Gillespie showed that sickling of the red cells was related to low oxygen.[11] In some individuals this change occurs at partial pressures of O
2 prevalent in the body, and produces anemia and other disorders, termed sickle-cell disease. In other persons sickling occurs only at very low O
2 partial pressures; these are asymptomatic sickle-cell trait carriers.
The association with kidney and lung infarcts was noted in 1931 by Yater and Mollari and Baird in 1934 respectively. The term sickle cell trait was coined by Samuel Diggs in Memphis in 1933 to distinguish heterozygotes from those with sickle cell anaemia. Diggs also reported the association with splenic fibrosis in 1935. The pathological mechanism of vaso-occlusion was proposed by Ham and Castle in 1940.
In 1946, E A Beet, a British medical officer stationed in Northern Rhodesia (Zimbabwe), observed that blood from malaria patients who had sickle cell trait had fewer malarial parasites than blood from patients without the trait and suggested that this might be a protective feature. In 1947 Beet published that the incidence of enlarged spleens in sickle cell patients was much lower than in non sickle cell and suggested that this was due to recurrent thromboses which resulted in fibrosis and shrinkage of the spleen. In 1949 Lehmann and Raper published a map of Uganda and showed that the presence of sickle cell anaemia correlated with the presence of malaria.[12] In 1950 Singer et al. noted the abrupt cessation of marrow activity that may occur and coined the term aplastic crisis. The role of parvovirus in aetiology of this condition was not recognised until 1981. P. Brain also while working in Northern Rhodesia confirmed the lower incidence of splenomegaly and suggested that while homozygotes for the sickle cell gene suffered from several problems heterozygotes might be protected against malaria.[13]
The modern phase of research on this disorder was initiated by the famous chemist Linus Pauling in 1949. Pauling postulated that the hemoglobin (Hb) in sickle-cell disease is abnormal; when deoxygenated it polymerizes into long, thin, helical rods that distort the red cell into a sickle shape. In his laboratory, electrophoretic studies showed that sickle-cell Hb (S) is indeed abnormal, having at physiological pH a lower negative charge than normal adult human Hb (A).[14] In sickle-cell trait carriers there is a nearly equal amount of HbA and HbS, whereas in persons with sickle-cell disease nearly all the Hb is of the S type, apart from a small amount of fetal Hb. These observations showed that most patients with sickle-cell disease are homozygous for the gene encoding HbS, while trait carriers are heterozygous for this gene. Persons inheriting a sickle-cell gene and another mutant at the same locus, e.g. a thalassemia gene, can also have a variant form of sickle-cell disease. Pauling also introduced the term "molecular disease", which, together with molecular medicine, has become widely used.
The next major advance was the discovery by Vernon Ingram in 1959 that HbS differs from HbA by only a single amino-acid substitution in the β-polypeptide chain (β6Glu → Val).[15] It was later established that this results from a substitution of thymine for adenine in the DNA codon (GAG → GTG). This was the first example in any species of the effects of a mutation on a protein. Sickle Hb induces the expression of heme oxygenase-1 in hematopoietic cells. Carbon monoxide, a byproduct of heme catabolism by heme oxygenase-1, prevents an accumulation of circulating free heme after Plasmodium infection, suppressing the pathogenesis of experimental cerebral malaria.[16] Other mechanisms, such as enhanced tolerance to disease mediated by HO-1 and reduced parasitic growth due to translocation of host micro-RNA into the parasite, have been described.[17]
Distribution of the sickle-cell gene
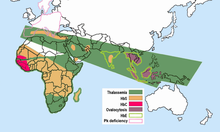
Since sickle-cell homozygotes are at a strong selective disadvantage, while protection against malaria favors the heterozygotes, it would be expected that high frequencies of the HbS gene would be found only in populations living in regions where malaria transmission is intense, or was so until the disease was eradicated. In a second study conducted in 1953 Allison showed that this was true in East Africa.[18] Frequencies of sickle-cell heterozygotes were 20-40% in malarious areas, whereas they were very low or zero in the highlands of Kenya, Uganda, and Tanzania. Later studies by many investigators filled in the picture.[19][20] High frequencies of the HbS gene are confined to a broad belt across Central Africa, but excluding most of Ethiopia and the East African highlands; this corresponds closely to areas of malaria transmission. Sickle-cell heterozygote frequencies up to 20% also occur in pockets of India and Greece that were formerly highly malarious. Tens of thousands of individuals have been studied, and high frequencies of abnormal hemoglobins have not been found in any population that was malaria free.
Independent origins of the sickle-cell gene

Two mutations found in nontranscribed sequences of DNA adjacent to the β-globin gene are so close to each other that the likelihood of crossover is very small. Restriction endonuclease digests of the β-globin gene cluster have shown five distinct patterns associated with the sickle-cell (GAG → GTG) mutation. Four are observed in Africa, the Bantu, Benin, Senegal and Cameroon types,[21] and a fifth type is found in the Indian subcontinent and Arabia.[22] The cited authors report that haplotype analysis in the β-globin region shows strong linkage disequilibrium over the distance indicated, which is evidence that the HbS mutation occurred independently at least five times. The high levels of AS in parts of Africa and India presumably resulted from independent selections occurring in different populations living in malarious environments. In summary, the demonstration that sickle-cell heterozygotes have some degree of protection against falciparum malaria was the first example of genetically controlled innate resistance to human malaria, as recognized by experts on inherited factors affecting human infectious diseases.[23] It was also the first demonstration of Darwinian selection in humans, as recognized by evolutionary biologists. [24]
Other abnormal hemoglobins
The frequencies of abnormal hemoglobins in different populations vary greatly, but some are undoubtedly polymorphic, having frequencies higher than expected by recurrent mutation. Four of these are α-thalassemia, which attains frequencies of 30% in parts of West Africa;[25] β-thalassemia, with frequencies up to 10% in parts of Italy; HbE (β26Glu → Lys), which attains frequencies up to 55% in Thailand and other Southeast Asian countries;[26] and HbC (β6Glu → Lys), which attains frequencies approaching 20% in northern Ghana and Burkina-Faso. All of these are in malarious areas, and there is evidence that the persons with α-thalassemia, HbC and HbE have some degree of protection against the parasite.[25][27][28] There is no longer doubt that malarial selection played a major role in the distribution of all these polymorphisms. An additional question is raised by the presence of polymorphisms for HbS and another Hb mutation in the sample population. Double heterozygotes for HbS and β-thalassemia, and for HbS and HbC, suffer from variant forms of sickle-cell disease, milder than SS but likely to reduce fitness before modern treatment was available. As predicted, these variant alleles tend to be mutually exclusive in populations. There is a negative correlation between frequencies of HbS and β-thalassemia in different parts of Greece and of HbS and HbC in West Africa.[29] Where there is no adverse interaction of mutations, as in the case of abnormal hemoglobins and G6PD deficiency, a positive correlation of these variant alleles in populations would be expected and is found.[29]
Thalassemias
It has long been known that a kind of anemia, termed thalassemia, has a high frequency in some Mediterranean populations, including Greeks and Southern Italians. The name is derived from the Greek words for sea (thalassa), meaning the Mediterranean sea, and blood (haima). Vernon Ingram again deserves the credit for explaining the genetic basis of different forms of thalassemia as an imbalance in the synthesis of the two polypeptide chains of Hb.[30] In the common Mediterranean variant, mutations decrease production of the β-chain (β thalassemia). In α-thalassemia, which is relatively frequent in Africa and several other countries, production of the α-chain of Hb is impaired, and there is relative over-production of the β-chain. Individuals homozygous for β-thalassemia have severe anemia and are unlikely to survive and reproduce, so selection against the gene is strong. Those homozygous for α thalassemia also suffer from anemia and there is some degree of selection against the gene.
Glucose-6-phosphate dehydrogenase deficiency

Glucose-6-phosphate dehydrogenase (G6PD) is an important enzyme in red cells, metabolizing glucose through the pentose phosphate pathway and maintaining a reducing environment. G6PD is present in all human cells but is particularly important to red blood cells. Since mature red blood cells lack nuclei and cytoplasmic RNA, they cannot synthesize new enzyme molecules to replace genetically abnormal or ageing ones. All proteins, including enzymes, have to last for the entire lifetime of the red blood cell, which is normally 120 days. In 1956 Alving and colleagues showed that in some African Americans the antimalarial drug primaquine induces hemolytic anemia, and that those individuals have an inherited deficiency of G6PD in erythrocytes.[31] G6PD deficiency is sex linked, and common in Mediterranean, African and other populations. In Mediterranean countries such individuals can develop a hemolytic diathesis (favism) after consuming fava beans. G6PD deficient persons are also sensitive to several drugs in addition to primaquine. G6PD deficiency is the most common enzyme deficiency in humans, estimated to affect some 400 million people.[32] There are many mutations at this locus, two of which attain frequencies of 20% or greater in African and Mediterranean populations; these are termed the A- and Med mutations.[33] Mutant varieties of G6PD can be more unstable than the naturally occurring enzyme, so that their activity declines more rapidly as red cells age.
Malaria in G6PD-deficient subjects
This question has been studied in isolated populations where antimalarial drugs were not used in Tanzania, East Africa[34] and in the Republic of the Gambia, West Africa, following children during the period when they are most susceptible to falciparum malaria.[35] In both cases parasite counts were significantly lower in G6PD-deficient persons than in those with normal red cell enzymes. The association has also been studied in individuals, which is possible because the enzyme deficiency is sex-linked and female heterozygotes are mosaics due to lyonization, where random inactivation of an X-chromosome in certain cells creates a population of G6PD deficient red blood cells coexisting with normal red blood cells. Malaria parasites were significantly more often observed in normal red cells than in enzyme-deficient cells.[36] An evolutionary genetic analysis of malarial selection on G6PD deficiency genes has been published by Tishkoff and Verelli.[33] The enzyme deficiency is common in many countries that are, or were formerly, malarious, but not elsewhere.
South-East Asian ovalocytosis
Ovalocytosis is an inherited condition in which erythrocytes have an oval instead of a round shape. In most populations ovalocytosis is rare, but South-East Asian ovalocytosis (SAO) occurs in as many as 15% of the indigenous people of Malaysia and of Papua New Guinea. Several abnormalities of SAO erythrocytes have been reported, including increased red cell rigidity and reduced expression of some red cell antigens.[38] SAO is caused by a mutation in the gene encoding the erythrocyte band 3 protein. There is a deletion of codons 400-408 in the gene, leading to a deletion of 9 amino-acids at the boundary between the cytoplasmic and transmembrane domains of band 3 protein.[39] Band 3 serves as the principal binding site for the membrane skeleton, a submembrane protein network composed of ankyrin, spectrin, actin, and band 4.1. Ovalocyte band 3 binds more tightly than normal band 3 to ankyrin, which connects the membrane skeleton to the band 3 anion transporter. These qualitative defects create a red blood cell membrane that is less tolerant of shear stress and more susceptible to permanent deformation.
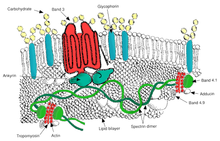
SAO is associated with protection against cerebral malaria in children because it reduces sequestration of erythrocytes parasitized by P. falciparum in the brain microvasculature.[40] Adhesion of P. falciparum-infected red blood cells to CD36 is enhanced by the cerebral malaria-protective SAO trait . Higher efficiency of sequestration via CD36 in SAO individuals could determine a different organ distribution of sequestered infected red blood cells. These provide a possible explanation for the selective advantage conferred by SAO against cerebral malaria.[41]
Resistance in South Asia
The lowest Himalayan Foothills and Inner Terai or Doon Valleys of Nepal and India are highly malarial due to a warm climate and marshes sustained during the dry season by groundwater percolating down from the higher hills. Malarial forests were intentionally maintained by the rulers of Nepal as a defensive measure. Humans attempting to live in this zone suffered much higher mortality than at higher elevations or below on the drier Gangetic Plain.
However, the Tharu people had lived in this zone long enough to evolve resistance via multiple genes. Medical studies among the Tharu and non-Tharu population of the Terai yielded the evidence that the prevalence of cases of residual malaria is nearly seven times lower among Tharus. The basis for their resistance to malaria is most likely a genetic factor. Endogamy along caste and ethnic lines appear to have confined these to the Tharu community.[42] Otherwise these genes probably would have become nearly universal in South Asia and beyond because of their considerable survival value and the apparent lack of negative effects comparable to Sickle Cell Anemia.
Duffy antigen receptor
The malaria parasite Plasmodium vivax is estimated to infect 75 million people annually. P. vivax has a wide distribution in tropical countries, but is absent or rare in a large region in West and Central Africa, as recently confirmed by PCR species typing.[43] This gap in distribution has been attributed to the lack of expression of the Duffy antigen receptor for chemokines (DARC) on the red cells of many sub-Saharan Africans. Duffy negative individuals are homozygous for a DARC allele, carrying a single nucleotide mutation (DARC 46 T → C), which impairs promoter activity by disrupting a binding site for the hGATA1 erythroid lineage transcription factor.[44] In widely cited in vitro and in vivo studies, Miller et al. reported that the Duffy blood group is the receptor for P. vivax and that the absence of the Duffy blood group on red cells is the resistance factor to P. vivax in persons of African descent.[45] This has become a well-known example of innate resistance to an infectious agent because of the absence of a receptor for the agent on target cells. However, observations have accumulated showing that the original report needs qualification. P. vivax can be transmitted in Squirrel monkeys (Saimiri boliviensis and S. sciureus), and Barnwell et al.[46] have obtained evidence that P. vivax enters Saimiri monkey red cells independently of the Duffy blood group, showing that P. vivax has an alternative pathway for invading these cells. The Duffy binding protein, the one and only invasion ligand for DARC, does not bind to Saimiri erythrocytes although these cells express DARC and obviously become infected with P. vivax.[47] The question is whether these observations are relevant to naturally occurring human transmission of P. vivax. Ryan et al. presented evidence for the transmission of P. vivax among a Duffy-negative population in Western Kenya.[48] Independently, Cavasini et al. have reported P. vivax infections in Duffy antigen-negative individuals from the Brazilian Amazon region.[49] P. vivax and Duffy antigen expression were identified by genotypic and other methods. A subsequent investigation in Madagascar has extended these observations.[50] The Malagasy people in this island have an admixture of Duffy-positive and Duffy-negative people of diverse ethnic backgrounds. At eight sentinel sites covering different parts of the island 72% of the populations were Duffy-negative, as shown by genotyping and flow cytometry. P. vivax positivity was found in 8.8% of 476 asymptomatic Duffy-negative people, and clinical P. vivax malaria was found in 17 such persons. Genotyping of polymorphic and microsatellite markers suggested that multiple P. vivax strains were invading the red cells of Duffy-negative people. The authors suggest that among Malagasy populations there are enough Duffy-positive people to maintain mosquito transmission and liver infection. From this internal source P. vivax variants can develop, using receptors other than Duffy to enter red cells and multiply. More recently, Duffy negative individuals infected with two different strains (VK247 and classic strains) of P. vivax were found in Angola and Equatorial Guinea; further, P. vivax infections were found both in humans and mosquitoes, which means that active transmission is occurring. This finding reinforces the idea that this parasite is able to use receptors other than Duffy to invade erythrocytes, which may have an enormous impact in P. vivax current distribution.[51] Because of these several reports from different parts of the world it is clear that some variants of P.vivax are being transmitted to humans who are not expressing DARC on their red cells. The frequency of such transmission is still unknown. Identification of the parasite and host molecules that allow Duffy-independent invasion of human erythrocytes is an important task for the future, because it may facilitate vaccine development.

P. vivax is clearly a less potent agent of natural selection that is P. falciparum. However, the morbidity of P. vivax is not negligible. For example, P. vivax infections induce a greater inflammatory response in the lungs than is observed in P. falciparum infections, and progressive alveolar capillary dysfunction is observed after the treatment of vivax malaria.[52] Epidemiological studies in the Amazonian region of Brazil have shown that the number and rate of hospital admissions for P. vivax infections have recently increased while those of P. falciparum have decreased.[53] Standard criteria for admission were used. The authors suggest that P. vivax infections in this region are becoming more severe. The distribution of Duffy negativity in Africa does not correlate precisely with that of P. vivax transmission.[43] Frequencies of Duffy negativity are as high in East Africa (above 80%), where the parasite is transmitted, as they are in West Africa, where it is not. In summary, P. vivax can bind to and invade human and nonhuman primate erythrocytes through a receptor or receptors other than DARC. However, DARC still appears to be a major receptor for human transmission of P. vivax. The potency of P. vivax as an agent of natural selection is unknown, and may vary from location to location. DARC negativity remains a good example of innate resistance to an infection, but it produces a relative and not an absolute resistance to P. vivax transmission.
Agent of natural selection
Natural selection was traditionally attributed to phenomena such as competition for resources or predation. There was no example of natural selection operating on a common gene in humans, in contrast to selection against rare deleterious mutations. After the Second World War an Italian group (E.Silvestroni, I.Bianco and G.Montalenti) developed methods for identifying ß-thalassemia heterozygotes in populations, and recorded their frequencies in different parts of Italy. In some regions heterozygote frequencies up to 10% were observed, and the strong geographic correspondence between the incidence of thalassemia and endemic malaria was noted, as documented by an Italian historian of science.[54] These researches "raised clearly the question of maintaining the frequency of a gene that, at that time, doomed homozygotes to death within the first two years of life". At an international meeting in Italy in 1949 J.B.S.Haldane gave an address on "Disease and Evolution".[55] In the ensuing discussion Montalenti presented information on the distribution of thalassemia in Italy, and acknowledged a suggestion from J.B.S. Haldane that thalassemic heterozygotes may be resistant to malaria.[56] Later in 1949 Haldane reiterated the same suggestion, with no reference to the Italian investigators.[57] Haldane is therefore widely regarded as the originator of the "malaria hypothesis". However, there have been suggestions that the role of Italian investigators in recognizing this correlation was insufficiently acknowledged,[58] and that opinion was also expressed by the Nobel prizewinning geneticist Joshua Lederberg.[59] Haldane’s general proposal that infections are important agents of natural selection was a timely reminder, but had a long parentage. It was first made by Alfred Russel Wallace, co-discoverer of natural selection as a cause of evolution,[60] and in the first half of the twentieth century several examples of genes conferring resistance to infections, and their implications for natural selection, were published, as noted by Lederberg.[59] Haldane conducted no research on abnormal hemoglobins or on malaria, and malaria was eradicated from Mediterranean countries after World War II, so the malaria hypothesis could not be validated on carriers of β-thalassemia.
Testing the malaria hypothesis

The first systematic investigation of the malaria hypothesis was conducted by Anthony Allison in East Africa in 1953. His initial study ascertained whether sickle-cell heterozygotes are protected against severe P. falciparum infections. This required working with children between four months and four years of age, when the morbidity and mortality from malaria is greatest. The study was done in Ugandan villages where antimalarial drugs were not used. Allison found that children in this age group carrying HbS had significantly lower malaria parasite counts than in those with HbA.[61] Severe morbidity and mortality in malaria were known to be correlated with high parasite counts. This observation has been confirmed many times in different parts of Africa, and potentially lethal manifestations of malaria (cerebral malaria and severe anemia) are rare in sickle-cell heterozygotes.[25][29] In the latter study the HbS carrier state was found to be negatively associated with all potentially lethal forms of P. falciparum malaria, whereas the negative associations of the carrier states of HbC and α-thalassemia were limited to cerebral malaria and severe anemia, respectively. These findings strongly suggest that, under conditions of intense P. falciparum transmission, young sickle-cell heterozygotes (AS) survive better than those with normal hemoglobin (AA), whereas sickle-cell homozygotes (SS) survive least well of all three genotypes.

Detailed study of a cohort of 1022 Kenyan children living near Lake Victoria, published in 2002, confirmed this prediction.[62] Many SS children still died before they attained one year of age. Between 2 and 16 months the mortality in AS children was found to be significantly lower than that in AA children. This well-controlled investigation shows the ongoing action of natural selection through disease in a human population. Analysis of genome-wide and fine-resolution association (GWA) is a powerful method for establishing the inheritance of resistance to infections and other diseases. Two independent preliminary analyses of GWA association with severe falciparum malaria in Africans have been carried out, one by the Malariagen Consortium in a Gambian population and the other by Rolf Horstmann (Bernhard Nocht Institute for Tropical Medicine, Hamburg) and his colleagues on a Ghanaian population. In both cases the only signal of association reaching genome-wide significance was with the HBB locus encoding the beta chain of hemoglobin, which is abnormal in HbS.[63] This does not imply that HbS is the only gene conferring innate resistance to falciparum malaria; there could be many such genes exerting more modest effects that are challenging to detect by GWA because of the low levels of linkage disequilibrium in African populations. However the same GWA association in two populations is powerful evidence that the single gene conferring strongest innate resistance to falciparum malaria is that encoding HbS.
Mechanisms of protection
The mechanisms by which erythrocytes containing abnormal hemoglobins, or are G6PD deficient, are partially protected against P. falciparum infections are not fully understood, although there has been no shortage of suggestions. During the peripheral blood stage of replication malaria parasites have a high rate of oxygen consumption[64] and ingest large amounts of hemoglobin.[65] It is likely that HbS in endocytic vesicles is deoxygenated, polymerizes and is poorly digested. In red cells containing abnormal hemoglobins, or which are G6PD deficient, oxygen radicals are produced, and malaria parasites induce additional oxidative stress.[66] This can result in changes in red cell membranes, including translocation of phosphatidylserine to their surface, followed by macrophage recognition and ingestion.[67] The authors suggest that this mechanism is likely to occur earlier in abnormal than in normal red cells, thereby restricting multiplication in the former. In addition, binding of parasitized sickle cells to endothelial cells is significantly decreased because of an altered display of P. falciparum erythrocyte membrane protein-1 (PfMP-1).[68] This protein is the parasite’s main cytoadherence ligand and virulence factor on the cell surface. During the late stages of parasite replication red cells are adherent to venous endothelium, and inhibiting this attachment could suppress replication.
Stimulation of adaptive immunity
One of the most interesting developments in biomedical science during the past few decades has been elucidation of mechanisms mediating innate immunity. One set of innate immune mechanisms is humoral, such as complement activation. Another set comprises pattern recognition receptors such as Toll-like receptors, which induce the production of interferons and other cytokines increasing resistance of cells such as monocytes to infections.[7] Cytokines produced during innate immune responses are among the activators of adaptive immune responses.[7] Antibodies exert additive or synergistic effects with mechanisms of innate immunity. Unstable HbS clusters Band-3, a major integral red cell protein;[66] antibodies recognize these clusters and accelerate their removal by phagocytic cells. Clustered Band 3 proteins with attached antibodies activate complement, and complement C3 fragments are opsonins recognized by the CR1 complement receptor on phagocytic cells.[69] A population study has shown that the protective effect of the sickle-cell trait against falciparum malaria involves the augmentation of adaptive as well as innate immune responses to the malaria parasite, illustrating the expected transition from innate to adaptive immunity.[70]
Fitnesses of different genotypes
The fitnesses of different genotypes in an African region where there is intense malarial selection were estimated by Anthony Allison in 1954.[71] In the Baamba population living in the Semliki Forest region in Western Uganda the sickle-cell heterozygote (AS) frequency is 40%, which means that the frequency of the sickle-cell gene is 0.255 and 6.5 of children born are SS homozygotes. If the frequency of the heterozygote is 0.40 the sickle-cell gene frequency (q) can be calculated from the Hardy-Weinberg equation 2q(1-q) = 0,40, whence q = 0.2555 and q2, the frequency of sickle-cell homozygotes, is 0.065. It is a reasonable assumption that until modern treatment was available three quarters of the SS homozygotes failed to reproduce. To balance this loss of sickle-cell genes, a mutation rate of 1:10.2 per gene per generation would be necessary. This is about 1000 times greater than mutation rates measured in Drosophila and other organisms and much higher than recorded for the sickle-cell locus in Africans.[72] To balance the polymorphism, Anthony Allison estimated that the fitness of the AS heterozygote would have to be 1.26 times than that of the normal homozygote. Later analyses of survival figures have given similar results, with some differences from site to site. In Gambians, it was estimated that AS heterozygotes have 90% protection against P. falciparum-associated severe anemia and cerebral malaria,[73] whereas in the Luo population of Kenya it was estimated that AS heterozygotes have 60% protection against severe malarial anemia.[62] These differences reflect the intensity of transmission of P. falciparum malaria from locality to locality and season to season, so fitness calculations will also vary. In many African populations the AS frequency is about 20%, and a fitness superiority over those with normal hemoglobin of the order of 10% is sufficient to produce a stable polymorphism.

Anthony Allison used the above fitness estimates to calculate the time that it would take for a newly mutated sickle-cell gene to increase in frequency in a population and attain a stable equilibrium. Under conditions of intense malarial selection the frequency of the sickle-cell gene would attain an equilibrium level in about 45 generations, slightly more than 1000 years. Under less intensive malarial selection the heterozygote advantage would be lower and a stable polymorphism with near 20% AS carriers (as commonly observed in Africa) would be attained in about 2,000 years. If malarial selection is relaxed, the frequency of the sickle-cell gene will fall exponentially. This has probably occurred in the African American population of the USA, but the rate of fall is uncertain because of the diverse and poorly documented African origin of the population as well as mixture in the USA with immigrants of other origins. Human genome sequencing can be applied not only to detect the effects of natural selection but also to obtain information about how recently it occurred. Sabeti and her colleagues have provided an appropriate framework.[74] First, haplotypes at a locus of interest are identified (core haplotypes). Then the age of each core haplotype is assessed by the decay of its association to alleles at various distances from the locus, as measured by extended haplotype homozygosity (EHH). Core haplotypes that have unusually high EHH as well as a high population prevalence reveal the presence of mutations that rose to prominence in the gene pool faster than expected by random drift. When this approach was applied to the G6PD locus, significant evidence of selection was found. A linkage-disequilibrium test was used to estimate the date of origin of the mutated G6PD gene conferring resistance, which gave a figure of about 2,500 years. Haplotype diversity and linkage disequilibrium using microsatellite data had previously been applied by Tishkoff and her colleagues to estimate the dates of origin of G6PD variants.[75] The African A- variant was estimated to have arisen within the past 3,840 to 11,760 years and the Med variant within the past 6,640 years. In summary, newly mutated abnormal hemoglobin and G6PD deficiency genes, arising in malarious environments, can quite rapidly become common and attain stable polymorphisms within 1,000 to 3,000 years, depending on the intensity of selection. Studies of genome variation and evolution of P. falciparum suggest that it originated within the last 3,200 to 7,700 years.[76] These dates coincide with the spread of agriculture within the last 10,000 years, which increased the density of populations, forest clearing, and urbanization near sunlit pools of water. Such conditions favor the breeding of Anopheles mosquitos and the transmission of malaria. The dramatic changes in human social organization since the onset of the Neolithic Age have had equally striking effects on the transmission of infectious diseases, and these, in turn, have had selective effects on human genes and left their signatures on the human genome.
Genetic factors influencing adaptive immunity
Humoral and cell-mediated immune responses limit malaria parasite multiplication, and many cytokines contribute to the pathogenesis of malaria as well as to the resolution of infections.[77] It is not surprising that genetic studies have identified several loci correlated with the severity of malaria.[78] For example, polymorphisms at the HLA loci, which encode proteins that participate in antigen presentation, influence the course of malaria. In West Africa an HLA class I antigen (HLA Bw53) and an HLA class II haplotype (DRB1*13OZ-DQB1*0501) are independently associated with protection against severe malaria.[73] However, HLA correlations vary, depending on the genetic constitution of the polymorphic malaria parasite, which differs in different geographic locations.[78]
Some early contributions on innate resistance to infections of vertebrates, including humans, are summarized in Table 1.
Table 1. Examples of Genetically Controlled Innate Resistance to Infectious Agents or Recognition of their Products
| Year of discovery | Pathogen | Mechanism of resistance | Authors |
|---|---|---|---|
| 1954 | P. falciparum | Sickle-cell heterozygote | Allison[61] |
| 1957 | Influenza virus | Interferon | Isaacs and Lindenmann[79] |
| 1976 | P.vivax | Non-expression of Duffy antigen on red cells | Miller et al.[45] |
| 1996 | Fungi | Toll receptor | Lemaitre[80] |
| 1998 | E.coli endotoxin | Toll-like receptor 4 | Poltorak[81] |
It is remarkable that two of the pioneering studies were on malaria. Type 1 interferons[79] and their mechanism of action have been analyzed in detail by genetic and other methods. The classical studies on the Toll receptor in Drosophila[80] were rapidly extended to Toll like receptors in mammals[81] and then to other pattern recognition receptors, which play important roles in innate immunity and its stimulation of adaptive immunity. The genetic control of innate and adaptive immunity is now a large and flourishing discipline. However, the early contributions on malaria remain as classical examples of innate resistance, which have stood the test of time.
References
- ^ López C, Saravia C, Gomez A, Hoebeke J, Patarroyo MA (2010). "Mechanisms of genetically-based resistance to malaria". Gene. 467 (1–2): 1–12. doi:10.1016/j.gene.2010.07.008. PMID 20655368.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kwiatkowski DP (2005). "How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria". Am J Hum Genet. 77 (2): 171–192. doi:10.1086/432519. PMC 1224522. PMID 16001361.
- ^ Hedrick PW (2011). "Population genetics of malaria resistance in humans". Heredity. 107 (4): 1–22. doi:10.1038/hdy.2011.16. PMC 3182497. PMID 21427751.
- ^ Anstee DJ (2010). "The relationship between blood groups and disease". Blood. 115 (23): 4635–4643. doi:10.1182/blood-2010-01-261859. PMID 20308598.
- ^ Fisher RA (1930). Full text of 1930 edition The Genetical Theory of Natural Selection "The Genetical Theory of Natural Selection". Cambridge University Press.
{{cite journal}}: Check|url=value (help) - ^ "CHU Hôpitaux de Rouen. Fréquence et origine des cas de paludisme". .chu-rouen.fr. Retrieved 2010-08-24.
- ^ a b c Uematsu S, Akira S (2007). "Toll-like receptors and Type I interferons". J Biol Chem. 282 (21): 15319–1523. doi:10.1074/jbc.R700009200. PMID 17395581.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lebby R (1846) Case of absence of the spleen. Southern J of Med Pharmacol 1: 481-483
- ^ Herrick JB (1910). "Peculiar elongated and sickle-shaped red blood corpuscles in a case of severe anemia". Arch Intern Med. 5: 517–521.; Reprinted in Herrick JB (2001). "Peculiar elongated and sickle-shaped red blood corpuscles in a case of severe anemia. 1910". Yale J Biol Med. 74 (3): 179–84. PMC 2588723. PMID 11501714.
- ^ Taliaffero WH, Huck JG (1923). "The Inheritance of Sickle-Cell Anaemia in Man". Genetics. 8 (6): 594–598. PMC 1200767. PMID 17246028.
- ^ Hahn EV, Gillespie EB (1927). "Sickle cell anemia. Report of a case greatly improved by splenectomy. Experimental study of sickle cell formation". Arch Intern Med. 39 (2): 233–254. doi:10.1001/archinte.1927.00130020072006.
- ^ Lehmann H, Raper AB (1949). "Distribution of sickle cell trait in Uganda, and its ethnological significance" (PDF). Nature. 164 (4168): 494–495. Bibcode:1949Natur.164..494L. doi:10.1038/164494a0. PMID 18140460.
- ^ Brain P (1952). "Sickle-cell Anaemia in Africa". Br Med J. 2: 880. PMC 2021738.
- ^ Pauling L, Itano HA, Singer SJ, Wells IC (1949). "Sickle cell anemia, a molecular disease". Science. 110 (2865): 543–548. Bibcode:1949Sci...110..543P. doi:10.1126/science.110.2865.543. PMID 15395398.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ingram VM. (1959). "Abnormal human haemoglobins. III. The chemical difference between normal and sickle cell haemoglobins". Biochim Biophys Acta. 36 (2): 402–411. doi:10.1016/0006-3002(59)90183-0. PMID 13852872.
- ^ Ferreira A, Marguti I, Bechmann I, Jeney V, Chora A, Palha NR, Rebelo S, Henri A, Beuzard Y, Soares MP (2011). "Sickle hemoglobin confers tolerance to Plasmodium infection" (PDF). Cell. 145 (3): 398–409. doi:10.1016/j.cell.2011.03.049. PMID 21529713.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gong L, Parikh S, Rosenthal PJ, Greenhouse B (2013). "Biochemical and immunological mechanisms by which sickle cell trait protects against malaria" (PDF). Malaria Journal. 12: 317. PMID 24025776.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Allison AC (1954). "The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria". Trans Roy Soc Trop Med Hyg. 48 (4): 312–318. doi:10.1016/0035-9203(54)90101-7. PMID 13187561.
- ^ a b Allison AC. (2009). "Genetic control of resistance to human malaria". Curr Opin Immunol. 21 (5): 499–505. doi:10.1016/j.coi.2009.04.001. PMID 19442502.
- ^ Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Williams TN, Weatherall DJ, Hay SI (2010). "Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis" (PDF). Nat Commun. 104 (8): 1–7. Bibcode:2010NatCo...1E.104P. doi:10.1038/ncomms1104. PMID 21045822.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lapouméroulie C, Dunda O, Ducrocq R, Trabuchet G, Mony-Lobé M, Bodo JM, Carnevale P, Labie D, Elion J, Krishnamoorthy R (1992). "A novel sickle cell mutation of yet another origin in Africa: the Cameroon type". Hum Genet. 89 (3): 333–337. PMID 1376298.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Labie D, Srinivas R, Dunda O, Dode C, Lapoumeroulie C, Devi V, Devi S, Ramasami K, Elion J, Ducrocq R; et al. (1989). "Haplotypes in tribal Indians bearing the sickle-gene: evidence for the unicentric origin of the βS mutation and the unicentric origin of tribal populations in India". Hum Biol. 61 (4): 479–491. PMID 2480325.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Alcaïs A, Abel L, Casanova JL (2009). "Human genetics of infectious diseases: between proof of principle and paradigm". J Clin Invest. 119 (9): 2506–2514. doi:10.1172/JCI38111. PMC 2735901. PMID 19729848.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Carroll SB, "A sickle-cell safari", Chapter 8 (pp. 148–165) in: Sean B. Carroll, Into the Jungle: Great Adventures in the Search for Evolution (San Francisco: Pearson Benjamin Cummings, 2009).
- ^ a b c May J, Evans JA, Timmann C, Ehmen C, Busch W, Thye T, Agbenyega T, Horstmann RD (2007). "Hemoglobin variants and disease manifestations in severe falciparum malaria". JAMA. 297 (20): 2220–2226. doi:10.1001/jama.297.20.2220. PMID 17519411.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Flatz G (1967). "Hemoglobin E: distribution and population dynamics". Humangenetik. 3 (3): 189–234. doi:10.1007/BF00273124. PMID 6074385.
- ^ Modiano D, Luoni G, Sirima BS, Simporé J, Verra F, Konaté A, Rastrelli E, Olivieri A, Calissano C, Paganotti GM, D'Urbano L, Sanou I, Sawadogo A, Modiano G, Coluzzi M (2001). "Haemoglobin C protects against clinical Plasmodium falciparum malaria". Nature. 414 (6861): 305–308. doi:10.1038/35104556. PMID 11713529.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hutagalung R, Wilairatana P, Looareesuwan S, Brittenham GM, Aikawa M, Gordeuk VR (1999). "Influence of hemoglobin E trait on the severity of Falciparum malaria". J Infect Dis. 179 (1): 283–286. doi:10.1086/314561. JSTOR 30117260. PMID 9841856.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Allison AC (1955). "Aspects of polymorphism in man". Cold Spring Harb Symp Quant Biol. 20: 239–251. PMID 13433567.
- ^ Ingram VM, Stretton AO (1959). "Genetic basis of the thalassaemia diseases". Nature. 184 (4703): 1903–1909. Bibcode:1959Natur.184.1903I. doi:10.1038/1841903a0. PMID 13852871.
- ^ Alving AS, Carson PE, Flanagan CL, Ickes CE (1956). "Enzymatic deficiency in primaquine-sensitive erythrocytes". Science. 124 (3220): 484–485. Bibcode:1956Sci...124..484C. doi:10.1126/science.124.3220.484-a. PMID 13360274.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Beutler E (2008). "Glucose-6-phosphate dehydrogenase deficiency: a historical perspective". Blood. 111 (1): 16–24. doi:10.1182/blood-2007-04-077412. PMID 18156501.
- ^ a b Tishkoff SA, Verelli BJ (2004). "G6PD deficiency and malarial resistance in humans: insights from evolutionary genetic analysis. In Evolutionary Aspects of Infectious Disease (Dronamraju K, ed)". Cambridge University Press.
- ^ Allison AC, Clyde DF (1961). "Malaria in African Children with Deficient Erythrocyte Glucose-6-phosphate Dehydrogenase". Br Med J. 1 (5236): 1346–1349. doi:10.1136/bmj.1.5236.1346. PMC 1954496. PMID 13682585.
- ^ Ruwende C, Khoo SC, Snow RW, Yates SNR, Kwiatkowski D, Gupta S, Warn P, Allsopp CEM, Gilbert SC, Peschu N, Newbold CI, Greenwood BM, Marsh K, Hill AVS (1995). "Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria". Nature. 376 (6537): 246–249. Bibcode:1995Natur.376..246R. doi:10.1038/376246a0. PMID 7617034.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Luzzatto L (1979). "Genetics of red cells and susceptibility to malaria" (PDF). Blood. 54 (5): 961–976. PMID 387115.
- ^ Hempelmann E, Götze O (1984). "Characterization of membrane proteins by polychromatic silver staining". Hoppe Seyler's Z Physiol Chem. 365: 241–242.
- ^ Jarolim P, Palek J, Amato D, Hassan K, Sapak P, Nurse GT, Rubin HL, Zhai S, Sahr KE, Liu SC (1991). "Deletion in erythrocyte band 3 gene in malaria-resistant Southeast Asian ovalocytosis" (PDF). Proc Natl Acad Sci U S A. 88 (24): 11022–11026. Bibcode:1991PNAS...8811022J. doi:10.1073/pnas.88.24.11022. PMC 53065. PMID 1722314.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Liu SC, Zhai S, Palek J, Golan DE, Amato D, Hassan K, Nurse GT, Babona D, Coetzer T, Jarolim P, Zaik M, Borwein S (1990). "Molecular defect of the band 3 protein in southeast Asian ovalocytosis". N Engl J Med. 323 (22): 1530–1538. doi:10.1056/NEJM199011293232205. PMID 2146504.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Allen SJ, O'Donnell A, Alexander ND, Mgone CS, Peto TE, Clegg JB, Alpers MP, Weatherall DJ (1999). "Prevention of cerebral malaria in children in Papua New Guinea by southeast Asian ovalocytosis band 3". Am J Trop Med Hyg. 60 (6): 1056–1060. PMID 10403343.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cortés A, Mellombo M, Mgone CS, Beck HP, Reeder JC, Cooke BM (2005). "Adhesion of Plasmodium falciparum-infected red blood cells to CD36 under flow is enhanced by the cerebral malaria-protective trait South-East Asian ovalocytosis". Mol Biochem Parasitol. 142 (2): 252–257. doi:10.1016/j.molbiopara.2005.03.016. PMID 15978955.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Terrenato L, Shrestha S, Dixit KA, Luzzatto L, Modiano G, Morpurgo G, Arese P (1988). "Decreased malaria morbidity in the Tharu people compared to sympatric populations in Nepal". Ann Trop Med Parasitol. 82 (1): 1–11. PMID 3041928.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Culleton RL, Mita T, Ndounga M, Unger H, Cravo PV, Paganotti GM, Takahashi N, Kaneko A, Eto H, Tinto H, Karema C, D'Alessandro U, do Rosário V, Kobayakawa T, Ntoumi F, Carter R, Tanabe K (2008). "Failure to detect Plasmodium vivax in West and Central Africa by PCR species typing". Malar J. 7 (1): 174–182. doi:10.1186/1475-2875-7-174. PMC 2546428. PMID 18783630.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Tournamille C, Colin Y, Cartron JP, Le Van Kim C (1995). "Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals". Nat Genet. 10 (2): 224–228. doi:10.1038/ng0695-224. PMID 7663520.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Miller LH, Mason SJ, Clyde DF, McGinniss MH (1976). "The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy". N Engl J Med. 295 (6): 302–4. doi:10.1056/NEJM197608052950602. PMID 778616.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Barnwell JW, Nichols ME, Rubinstein P (1989). "In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax". J Exp Med. 169 (5): 1795–802. doi:10.1084/jem.169.5.1795. PMC 2189319. PMID 2469769.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wertheimer SP, Barnwell JW (1989). "Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein". Exp Parasitol. 69 (4): 340–350. doi:10.1016/0014-4894(89)90083-0. PMID 2680568.
- ^ Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, Koros J, Owour B, Luckhart S, Wirtz RA, Barnwell JW, Rosenberg R (2006). "Evidence for transmission of Plasmodium vivax among a duffy antigen negative population in Western Kenya". Am J Trop Med Hyg. 75 (4): 575–581. PMID 17038676.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cavasini CE, de Mattos LC, Couto AA, Couto VS, Gollino Y, Moretti LJ, Bonini-Domingos CR, Rossit AR, Castilho L, Machado RL (2007). "Duffy blood group gene polymorphisms among malaria vivax patients in four areas of the Brazilian Amazon region". Malar J. 6 (1): 167. doi:10.1186/1475-2875-6-167. PMC 2244634. PMID 18093292.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod JF, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA (2010). "Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people". Proc Natl Acad Sci U S A. 107 (13): 5967–71. Bibcode:2010PNAS..107.5967M. doi:10.1073/pnas.0912496107. PMC 2851935. PMID 20231434.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosário VE, Benito A, Berzosa P, Arez AP (2011). Franco-Paredes, Carlos (ed.). "Duffy Negative Antigen Is No Longer a Barrier to Plasmodium vivax – Molecular Evidences from the African West Coast (Angola and Equatorial Guinea)". PLoS Negl Trop Dis. 5 (e1192): e1192. doi:10.1371/journal.pntd.0001192. PMC 3119644. PMID 21713024.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Anstey NM, Handojo T, Pain MC, Kenangalem E, Tjitra E, Price RN, Maguire GP (2007). "Lung Injury in Vivax Malaria: Pathophysiological Evidence for Pulmonary Vascular Sequestration and Posttreatment Alveolar-Capillary Inflammation". J Infect Dis. 195 (4): 589–596. doi:10.1086/510756. PMC 2532499. PMID 17230420.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Santos-Ciminera PD, Roberts DR, Alecrim MGC, Costa MRF, Quinnan GV Jr (2007). "Malaria Diagnosis and Hospitalization Trends, Brazil" (PDF). Emerg Infect Dis. 13 (10): 1597–1600. PMC 2851511. PMID 18258018.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Canali S (2008). "Researches on thalassemia and malaria in Italy and the origins of the "Haldane hypothesis"". Med Secoli. 20 (3): 827–846. PMID 19848219.
- ^ Haldane JBS (1949). "Disease and evolution". Ric Sci. 19 (Suppl A): 68–76.
- ^ Montalenti G (1949). "Comment on Haldane, JBS. Disease and evolution". Ric Sci. 19 (Suppl A): 333–334.
- ^ Haldane JBS (1949). "The rate of mutation of human genes". Proc Int Congr Genet Hered. 35 (Suppl): 267–273.
- ^ Ceppellini R (1955). "Discussion of "Aspects of Polymorphism in Man"". Cold Spring Harbor Symp Quant Biol. 20: 251–255.
- ^ a b Lederberg J (1999). "J. B. S. Haldane (1949) on infectious disease and evolution". Genetics. 153 (1): 1–3. PMC 1460735. PMID 10471694.
- ^ Williams-Ellis Amabel, Darwin's Moon: a Biography of Alfred Russel Wallace (London: Blackie, 1966).
- ^ a b Allison AC (1954). "Protection Afforded by Sickle-cell Trait Against Subtertian Malarial Infection" (PDF). Br Med J. 1 (4857): 290–294. doi:10.1136/bmj.1.4857.290. PMC 2093356. PMID 13115700.
- ^ a b c Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V (2002). "Protective effects of the sickle cell gene against malaria morbidity and mortality" (PDF). Lancet. 359 (9314): 1311–1312. doi:10.1016/S0140-6736(02)08273-9. PMID 11965279.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jallow, M; Teo, YY; Small, KS; Rockett, KA; Deloukas, P; Clark, TG; Kivinen, K; Bojang, KA; Conway, DJ (2009). "Genome-wide and fine-resolution association analysis of malaria in West Africa". Nat Genet. 41 (6): 657–665. doi:10.1038/ng.388. PMC 2889040. PMID 19465909.
- ^ Vaidya AB, Mather MW (2009). "Mitochondrial evolution and functions in malaria parasites". Annu Rev Microbiol. 63: 249–267. doi:10.1146/annurev.micro.091208.073424. PMID 19575561.
- ^ Elliott DA, McIntosh MT, Hosgood HD 3rd, Chen S, Zhang G, Baevova P, Joiner KA (2008). "Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum" (PDF). Proc Natl Acad Sci U S A. 105 (7): 2463–2468. Bibcode:2008PNAS..105.2463E. doi:10.1073/pnas.0711067105. PMC 2268159. PMID 18263733.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ a b Kuross SA, Rank BH, Hebbel RP (1988). "Excess heme in sickle erythrocyte inside-out membranes: possible role in thiol oxidation" (PDF). Blood. 71 (4): 876–882. PMID 3355895.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Föller M, Bobbala D, Koka S, Huber SM, Gulbins E, Lang F (2009). "Suicide for survival--death of infected erythrocytes as a host mechanism to survive malaria" (PDF). Cell Physiol Biochem. 24 (3–4): 133–140. doi:10.1159/000233238. PMID 19710527.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cholera R, Brittain NJ, Gillrie MR, Lopera-Mesa TM, Diakité SA, Arie T, Krause MA, Guindo A, Tubman A, Fujioka H, Diallo DA, Doumbo OK, Ho M, Wellems TE, Fairhurst RM (2008). "Impaired cytoadherence of Plasmodium falciparum-infected erythrocytes containing sickle hemoglobin" (PDF). Proc Natl Acad Sci U S A. 105 (3): 991–996. Bibcode:2008PNAS..105..991C. doi:10.1073/pnas.0711401105. PMC 2242681. PMID 18192399.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Arese P, Turrini F, Schwarzer E (2005). "Band 3/complement-mediated recognition and removal of normally senescent and pathological human erythrocytes" (PDF). Cell Physiol Biochem. 16 (4–6): 133–146. doi:10.1159/000089839. PMID 16301814.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Williams TN, Mwangi TW, Roberts DJ, Alexander ND, Weatherall DJ, Wambua S, Kortok M, Snow RW, Marsh K (2005). "An Immune Basis for Malaria Protection by the Sickle Cell Trait". PLoS Med. 2 (5): e128. doi:10.1371/journal.pmed.0020128. PMC 1140945. PMID 15916466.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Allison AC (1954). "Notes on sickle-cell polymorphism". Ann Hum Genet. 19: 39–57. doi:10.1111/j.1469-1809.1954.tb01261.x.
- ^ Vandepitte JM, Zuelzer WW, Neel JV, Colaert J (1955). "Evidence concerning the inadequacy of mutation as an explanation of the frequency of the sickle cell gene in the Belgian Congo". Blood. 10 (4): 341–350. PMID 14363315.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM (1991). "Common west African HLA antigens are associated with protection from severe malaria". Nature. 352 (6336): 595–600. Bibcode:1991Natur.352..595H. doi:10.1038/352595a0. PMID 1865923.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES (2002). "Detecting recent positive selection in the human genome from haplotype structure". Nature. 419 (6909): 832–837. Bibcode:2002Natur.419..832S. doi:10.1038/nature01140. PMID 12397357.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, Piro A, Stoneking M, Tagarelli A, Tagarelli G, Touma EH, Williams SM, Clark AG (2001). "Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance". Science. 293 (5529): 455–462. doi:10.1126/science.1061573. PMID 11423617.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M (2007). "Genome variation and evolution of the malaria parasite Plasmodium falciparum". Nat Genet. 39 (1): 120–125. doi:10.1038/ng1931. PMC 2663918. PMID 17159978.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schofield L, Grau GE (2005). "Immunological processes in malaria pathogenesis". Nat Rev Immunol. 5 (9): 722–735. doi:10.1038/nri1686. PMID 16138104.
- ^ a b Frodsham AJ, Hill AV (2004). "Genetics of infectious diseases". Hum Mol Genet. 13 Spec No 2: R187–R194. doi:10.1093/hmg/ddh225. PMID 15358724.
- ^ a b Isaacs A, Lindenmann J (1957). "Virus interference. I. The interferon" (PDF). Proc R Soc Lond B Biol Sci. 147 (927): 258–267. Bibcode:1957RSPSB.147..258I. doi:10.1098/rspb.1957.0048. PMID 13465720.
- ^ a b Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA (1996). "The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults". Cell. 86 (6): 973–983. doi:10.1016/S0092-8674(00)80172-5. PMID 8808632.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B (1998). "Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene". Science. 282 (5396): 2085–2088. Bibcode:1998Sci...282.2085P. doi:10.1126/science.282.5396.2085. PMID 9851930.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Further reading
- Krishna R Dronamraju, Paolo Arese (2006) Malaria: Genetic and Evolutionary Aspects, Springer; Berlin, ISBN 0-387-28294-7 / ISBN 978-0-387-28294-7
External links
- Favism
- Glucose-6-Phosphate Dehydrogenase deficiency an estimated 400 million people worldwide are affected by this enzymopathy
- Hemoglobinopathies
- Malaria and the Red Cell
- Malaria lectures
- Sickle Cell Disease
- Thalassemia
- World Malaria Report 2009 More than one-third of the 108 malarious countries documented reduction in malaria cases of app 50% in 2008 compared to 2000
