Stop codon
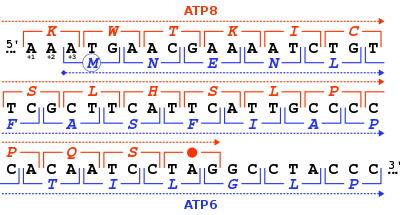
In molecular biology, a stop codon (or termination codon) is a codon (nucleotide triplet within messenger RNA) that signals the termination of the translation process of the current protein.[1] Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.
While start codons need nearby sequences or initiation factors to start translation, a stop codon alone is sufficient to initiate termination.
Properties[edit]
Standard codons[edit]
In the standard genetic code, there are three different termination codons:
| Codon | Standard code (Translation table 1) |
Name | ||
|---|---|---|---|---|
| DNA | RNA | |||
TAG |
UAG
|
STOP = Ter (*)
|
"amber" | |
TAA |
UAA
|
STOP = Ter (*)
|
"ochre" | |
TGA |
UGA
|
STOP = Ter (*)
|
"opal" (or "umber") | |
Alternative stop codons[edit]
There are variations on the standard genetic code, and alternative stop codons have been found in the mitochondrial genomes of vertebrates,[2] Scenedesmus obliquus,[3] and Thraustochytrium.[4]
| Genetic code | Translation table |
Codon | Translation with this code |
Standard translation | ||||
|---|---|---|---|---|---|---|---|---|
| DNA | RNA | |||||||
| Vertebrate mitochondrial | 2 | AGA |
AGA |
STOP = Ter (*) |
Arg (R)
| |||
AGG |
AGG |
STOP = Ter (*) |
Arg (R)
| |||||
| Scenedesmus obliquus mitochondrial | 22 | TCA |
UCA |
STOP = Ter (*) |
Ser (S)
| |||
| Thraustochytrium mitochondrial | 23 | TTA |
UUA |
STOP = Ter (*) |
Leu (L)
| |||
| Amino-acid biochemical properties | Nonpolar | Polar | Basic | Acidic | Termination: stop codon |
Reassigned stop codons[edit]
The nuclear genetic code is flexible as illustrated by variant genetic codes that reassign standard stop codons to amino acids.[5]
| Genetic code | Translation table |
Codon | Conditional translation |
Standard translation | ||||
|---|---|---|---|---|---|---|---|---|
| DNA | RNA | |||||||
| Karyorelict nuclear | 27 | TGA |
UGA |
Ter (*) |
or | Trp (W) |
Ter (*)
| |
| Condylostoma nuclear | 28 | TAA |
UAA |
Ter (*) |
or | Gln (Q) |
Ter (*)
| |
TAG |
UAG |
Ter (*) |
or | Gln (Q) |
Ter (*)
| |||
TGA |
UGA |
Ter (*) |
or | Trp (W) |
Ter (*)
| |||
| Blastocrithidia nuclear | 31 | TAA |
UAA |
Ter (*) |
or | Glu (E) |
Ter (*)
| |
TAG |
UAG |
Ter (*) |
or | Glu (E) |
Ter (*)
| |||
Translation[edit]
In 1986, convincing evidence was provided that selenocysteine (Sec) was incorporated co-translationally. Moreover, the codon partially directing its incorporation in the polypeptide chain was identified as UGA also known as the opal termination codon.[6] Different mechanisms for overriding the termination function of this codon have been identified in prokaryotes and in eukaryotes.[7] A particular difference between these kingdoms is that cis elements seem restricted to the neighborhood of the UAG codon in prokaryotes while in eukaryotes this restriction is not present. Instead such locations seem disfavored albeit not prohibited. [8]
In 2003, a landmark paper described the identification of all known selenoproteins in humans: 25 in total.[9] Similar analyses have been run for other organisms.
The UAG codon can translate into pyrrolysine (Pyl) in a similar manner.
Genomic distribution[edit]
Distribution of stop codons within the genome of an organism is non-random and can correlate with GC-content.[10][11] For example, the E. coli K-12 genome contains 2705 TAA (63%), 1257 TGA (29%), and 326 TAG (8%) stop codons (GC content 50.8%).[12] Also the substrates for the stop codons release factor 1 or release factor 2 are strongly correlated to the abundance of stop codons.[11] Large scale study of bacteria with a broad range of GC-contents shows that while the frequency of occurrence of TAA is negatively correlated to the GC-content and the frequency of occurrence of TGA is positively correlated to the GC-content, the frequency of occurrence of the TAG stop codon, which is often the minimally used stop codon in a genome, is not influenced by the GC-content.[13]
Recognition[edit]
Recognition of stop codons in bacteria have been associated with the so-called 'tripeptide anticodon',[14] a highly conserved amino acid motif in RF1 (PxT) and RF2 (SPF). Even though this is supported by structural studies, it was shown that the tripeptide anticodon hypothesis is an oversimplification.[15]
Nomenclature[edit]
Stop codons were historically given many different names, as they each corresponded to a distinct class of mutants that all behaved in a similar manner. These mutants were first isolated within bacteriophages (T4 and lambda), viruses that infect the bacteria Escherichia coli. Mutations in viral genes weakened their infectious ability, sometimes creating viruses that were able to infect and grow within only certain varieties of E. coli.
amber mutations (UAG)[edit]
They were the first set of nonsense mutations to be discovered, isolated by Richard H. Epstein and Charles Steinberg and named after their friend and graduate Caltech student Harris Bernstein, whose last name means "amber" in German (cf. Bernstein).[16][17]
Viruses with amber mutations are characterized by their ability to infect only certain strains of bacteria, known as amber suppressors. These bacteria carry their own mutation that allows a recovery of function in the mutant viruses. For example, a mutation in the tRNA that recognizes the amber stop codon allows translation to "read through" the codon and produce a full-length protein, thereby recovering the normal form of the protein and "suppressing" the amber mutation.[18] Thus, amber mutants are an entire class of virus mutants that can grow in bacteria that contain amber suppressor mutations. Similar suppressors are known for ochre and opal stop codons as well.
tRNA molecules carrying unnatural aminoacids have been designed to recognize the amber stop codon in bacterial RNA. This technology allows for incorporation of orthogonal aminoacids (such as p-azidophenylalanine) at specific locations of the target protein.
ochre mutations (UAA)[edit]
It was the second stop codon mutation to be discovered. Reminiscent of the usual yellow-orange-brown color associated with amber, this second stop codon was given the name of "ochre", an orange-reddish-brown mineral pigment.[17]
Ochre mutant viruses had a property similar to amber mutants in that they recovered infectious ability within certain suppressor strains of bacteria. The set of ochre suppressors was distinct from amber suppressors, so ochre mutants were inferred to correspond to a different nucleotide triplet. Through a series of mutation experiments comparing these mutants with each other and other known amino acid codons, Sydney Brenner concluded that the amber and ochre mutations corresponded to the nucleotide triplets "UAG" and "UAA".[19]
opal or umber mutations (UGA)[edit]
The third and last stop codon in the standard genetic code was discovered soon after, and corresponds to the nucleotide triplet "UGA".[20]
To continue matching with the theme of colored minerals, the third nonsense codon came to be known as "opal", which is a type of silica showing a variety of colors.[17] Nonsense mutations that created this premature stop codon were later called opal mutations or umber mutations.
Mutations and disease[edit]
Nonsense[edit]
Nonsense mutations are changes in DNA sequence that introduce a premature stop codon, causing any resulting protein to be abnormally shortened. This often causes a loss of function in the protein, as critical parts of the amino acid chain are no longer assembled. Because of this terminology, stop codons have also been referred to as nonsense codons.
Nonstop[edit]
A nonstop mutation, also called a stop-loss variant, is a point mutation that occurs within a stop codon. Nonstop mutations cause the continued translation of an mRNA strand into what should be an untranslated region. Most polypeptides resulting from a gene with a nonstop mutation lose their function due to their extreme length and the impact on normal folding. Nonstop mutations differ from nonsense mutations in that they do not create a stop codon but, instead, delete one. Nonstop mutations also differ from missense mutations, which are point mutations where a single nucleotide is changed to cause replacement by a different amino acid. Nonstop mutations have been linked with many inherited diseases including endocrine disorders,[21] eye disease,[22] and neurodevelopmental disorders.[23][24]
Hidden stops[edit]
Hidden stops are non-stop codons that would be read as stop codons if they were frameshifted +1 or −1. These prematurely terminate translation if the corresponding frame-shift (such as due to a ribosomal RNA slip) occurs before the hidden stop. It is hypothesised that this decreases resource wastage on nonfunctional proteins and the production of potential cytotoxins. Researchers at Louisiana State University propose the ambush hypothesis, that hidden stops are selected for. Codons that can form hidden stops are used in genomes more frequently compared to synonymous codons that would otherwise code for the same amino acid. Unstable rRNA in an organism correlates with a higher frequency of hidden stops.[25] However, this hypothesis could not be validated with a larger data set.[26]
Stop-codons and hidden stops together are collectively referred as stop-signals. Researchers at University of Memphis found that the ratios of the stop-signals on the three reading frames of a genome (referred to as translation stop-signals ratio or TSSR) of genetically related bacteria, despite their great differences in gene contents, are much alike. This nearly identical genomic-TSSR value of genetically related bacteria may suggest that bacterial genome expansion is limited by their unique stop-signals bias of that bacterial species.[27]
Translational readthrough[edit]
Stop codon suppression or translational readthrough occurs when in translation a stop codon is interpreted as a sense codon, that is, when a (standard) amino acid is 'encoded' by the stop codon. Mutated tRNAs can be the cause of readthrough, but also certain nucleotide motifs close to the stop codon. Translational readthrough is very common in viruses and bacteria, and has also been found as a gene regulatory principle in humans, yeasts, bacteria and drosophila.[28][29] This kind of endogenous translational readthrough constitutes a variation of the genetic code, because a stop codon codes for an amino acid. In the case of human malate dehydrogenase, the stop codon is read through with a frequency of about 4%.[30] The amino acid inserted at the stop codon depends on the identity of the stop codon itself: Gln, Tyr, and Lys have been found for the UAA and UAG codons, while Cys, Trp, and Arg for the UGA codon have been identified by mass spectrometry.[31] Extent of readthrough in mammals have widely variable extents, and can broadly diversify the proteome and affect cancer progression.[32]
Use as a watermark[edit]
In 2010, when Craig Venter unveiled the first fully functioning, reproducing cell controlled by synthetic DNA he described how his team used frequent stop codons to create watermarks in RNA and DNA to help confirm the results were indeed synthetic (and not contaminated or otherwise), using it to encode authors' names and website addresses.[33]
See also[edit]
References[edit]
- ^ Griffiths AJF, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM (2000). "Chapter 10 (Molecular Biology of Gene Function): Genetic code: Stop codons". An Introduction to Genetic Analysis. W.H. Freeman and Company.
- ^ Barrell, B. G.; Bankier, A. T.; Drouin, J. (1979-11-08). "A different genetic code in human mitochondria". Nature. 282 (5735): 189–194. Bibcode:1979Natur.282..189B. doi:10.1038/282189a0. ISSN 0028-0836. PMID 226894. S2CID 4335828.
- ^ A. M. Nedelcu, R. W. Lee, G. Lemieux, M. W. Gray, G. Burger (June 2000). "The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome". Genome Research. 10 (6): 819–831. doi:10.1101/gr.10.6.819. PMC 310893. PMID 10854413.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wideman, Jeremy G.; Monier, Adam; Rodríguez-Martínez, Raquel; Leonard, Guy; Cook, Emily; Poirier, Camille; Maguire, Finlay; Milner, David S.; Irwin, Nicholas A. T.; Moore, Karen; Santoro, Alyson E. (2019-11-25). "Unexpected mitochondrial genome diversity revealed by targeted single-cell genomics of heterotrophic flagellated protists". Nature Microbiology. 5 (1): 154–165. doi:10.1038/s41564-019-0605-4. hdl:10871/39819. ISSN 2058-5276. PMID 31768028. S2CID 208279678.
- ^ Swart, Estienne Carl; Serra, Valentina; Petroni, Giulio; Nowacki, Mariusz (2016). "Genetic Codes with No Dedicated Stop Codon: Context-Dependent Translation Termination". Cell. 166 (3): 691–702. doi:10.1016/j.cell.2016.06.020. PMC 4967479. PMID 27426948.
- ^ Zinoni, F; Birkmann, A; Stadtman, T; Böck, A (1986). "Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli". Proceedings of the National Academy of Sciences. 83 (13): 4650–4654. Bibcode:1986PNAS...83.4650Z. doi:10.1073/pnas.83.13.4650. PMC 323799. PMID 2941757.
- ^ Böck, A (2013). "Selenoprotein Synthesis". Encyclopedia of Biological Chemistry. pp. 210–213. doi:10.1016/B978-0-12-378630-2.00025-6. ISBN 9780123786319. Retrieved 23 August 2021.
- ^ Mix, H; Lobanov, A; Gladyshev, V (2007). "SECIS elements in the coding regions of selenoprotein transcripts are functional in higher eukaryotes". Nucleic Acids Research. 35 (2): 414–423. doi:10.1093/nar/gkl1060. PMC 1802603. PMID 17169995.
- ^ Kryukov, G; Gladyshev, V (2003). "Characterization of mammalian selenoproteomes". Science. 300 (5624): 1439–1443. Bibcode:2003Sci...300.1439K. doi:10.1126/science.1083516. PMID 12775843. S2CID 10363908.
- ^ Povolotskaya IS, Kondrashov FA, Ledda A, Vlasov PK (2012). "Stop codons in bacteria are not selectively equivalent". Biology Direct. 7: 30. doi:10.1186/1745-6150-7-30. PMC 3549826. PMID 22974057.
- ^ a b Korkmaz, Gürkan; Holm, Mikael; Wiens, Tobias; Sanyal, Suparna (2014). "Comprehensive Analysis of Stop Codon Usage in Bacteria and Its Correlation with Release Factor Abundance". The Journal of Biological Chemistry. 289 (44): 775–806. doi:10.1074/jbc.M114.606632. PMC 4215218. PMID 25217634.
- ^ "Escherichia coli str. K-12 substr. MG1655, complete genome [Genbank Accession Number: U00096]". GenBank. NCBI. Retrieved 2013-01-27.
- ^ Wong, Tit-Yee; Fernandes, Sanjit; Sankhon, Naby; Leong, Patrick P; Kuo, Jimmy; Liu, Jong-Kang (2008). "Role of Premature Stop Codons in Bacterial Evolution". Journal of Bacteriology. 190 (20): 6718–6725. doi:10.1128/JB.00682-08. PMC 2566208. PMID 18708500.
- ^ Ito, Koichi; Uno, Makiko; Nakamura, Yoshikazu (1999). "A tripeptide 'anticodon' deciphers stop codons in messenger RNA". Nature. 403 (6770): 680–684. doi:10.1038/35001115. PMID 10688208. S2CID 4331695.
- ^ Korkmaz, Gürkan; Sanyal, Suparna (2017). "R213I mutation in release factor 2 (RF2) is one step forward for engineering an omnipotent release factor in bacteria Escherichia coli". Journal of Biological Chemistry. 292 (36): 15134–15142. doi:10.1074/jbc.M117.785238. PMC 5592688. PMID 28743745.
- ^ Stahl FW (1995). "The amber mutants of phage T4". Genetics. 141 (2): 439–442. doi:10.1093/genetics/141.2.439. PMC 1206745. PMID 8647382.
- ^ a b c Lewin, Benjamin; Krebs, Jocelyn E.; Goldstein, Elliott S.; Kilpatrick, Stephen T. (2011-04-18). Lewin's Essential GENES. Jones & Bartlett Publishers. ISBN 978-1-4496-4380-5.
- ^ Robin Cook. "Amber, Ocher, and Opal Mutations Summary". World of Genetics. Gale.
- ^ Brenner, S.; Stretton, A. O. W.; Kaplan, S. (1965). "Genetic Code: The 'Nonsense' Triplets for Chain Termination and their Suppression". Nature. 206 (4988): 994–8. Bibcode:1965Natur.206..994B. doi:10.1038/206994a0. PMID 5320272. S2CID 28502898.
- ^ Brenner, S.; Barnett, L.; Katz, E. R.; Crick, F. H. C. (1967). "UGA: A Third Nonsense Triplet in the Genetic Code". Nature. 213 (5075): 449–50. Bibcode:1967Natur.213..449B. doi:10.1038/213449a0. PMID 6032223. S2CID 4211867.
- ^ Pang S.; Wang W.; et al. (2002). "A novel nonstop mutation in the stop codon and a novel missense mutation in the type II 3beta-hydroxysteroid dehydrogenase (3beta-HSD) gene causing, respectively, nonclassic and classic 3beta-HSD deficiency congenital adrenal hyperplasia". J Clin Endocrinol Metab. 87 (6): 2556–63. doi:10.1210/jcem.87.6.8559. PMID 12050213.
- ^ Doucette, L.; et al. (2011). "A novel, non-stop mutation in FOXE3 causes an autosomal dominant form of variable anterior segment dysgenesis including Peters anomaly". European Journal of Human Genetics. 19 (3): 293–299. doi:10.1038/ejhg.2010.210. PMC 3062009. PMID 21150893.
- ^ Torres-Torronteras, J.; Rodriguez-Palmero, A.; et al. (2011). "A novel nonstop mutation in TYMP does not induce nonstop mRNA decay in a MNGIE patient with severe neuropathy" (PDF). Hum. Mutat. 32 (4): E2061–E2068. doi:10.1002/humu.21447. PMID 21412940. S2CID 24446773.
- ^ Spaull, R; Steel, D; Barwick, K; Prabhakar, P; Wakeling, E; Kurian, MA (2022-07-23). "STXBP1 Stop-Loss Mutation Associated with Complex Early Onset Movement Disorder without Epilepsy". Movement Disorders Clinical Practice. 9 (6): 837–840. doi:10.1002/mdc3.13509. ISSN 2330-1619. PMC 9346254. PMID 35937496.
- ^ Seligmann, Hervé; Pollock, David D. (2004). "The Ambush Hypothesis: Hidden Stop Codons Prevent Off-Frame Gene Reading". DNA and Cell Biology. 23 (10): 701–5. doi:10.1089/1044549042476910. PMID 15585128.
- ^ Cavalcanti, Andre; Chang, Charlotte H.; Morgens, David W. (2013). "Ambushing the ambush hypothesis: predicting and evaluating off-frame codon frequencies in Prokaryotic Genomes". BMC Genomics. 14 (418): 1–8. doi:10.1186/1471-2164-14-418. PMC 3700767. PMID 23799949.
- ^ Wong, Tit-Yee; Schwartzbach, Steve (2015). "Protein mis-termination initiates genetic diseases, cancers, and restricts bacterial genome expansion". Journal of Environmental Science and Health, Part C. 33 (3): 255–85. doi:10.1080/10590501.2015.1053461. PMID 26087060. S2CID 20380447.
- ^ Namy O, Rousset JP, Napthine S, Brierley I (2004). "Reprogrammed genetic decoding in cellular gene expression". Molecular Cell. 13 (2): 157–68. doi:10.1016/S1097-2765(04)00031-0. PMID 14759362.
- ^ Schueren F, Lingner T, George R, Hofhuis J, Gartner J, Thoms S (2014). "Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals". eLife. 3: e03640. doi:10.7554/eLife.03640. PMC 4359377. PMID 25247702.
- ^ Hofhuis J, Schueren F, Nötzel C, Lingner T, Gärtner J, Jahn O, Thoms S (2016). "The functional readthrough extension of malate dehydrogenase reveals a modification of the genetic code". Open Biol. 6 (11): 160246. doi:10.1098/rsob.160246. PMC 5133446. PMID 27881739.
- ^ Blanchet S, Cornu D, Argentini M, Namy O (2014). "New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae". Nucleic Acids Res. 42 (15): 10061–72. doi:10.1093/nar/gku663. PMC 4150775. PMID 25056309.
- ^ Ghosh, Souvik; Guimaraes, Joao C; Lanzafame, Manuela; Schmidt, Alexander; Syed, Afzal Pasha; Dimitriades, Beatrice; Börsch, Anastasiya; Ghosh, Shreemoyee; Mittal, Nitish; Montavon, Thomas; Correia, Ana Luisa; Danner, Johannes; Meister, Gunter; Terracciano, Luigi M; Pfeffer, Sébastien; Piscuoglio, Salvatore; Zavolan, Mihaela (15 September 2020). "Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation". The EMBO Journal. 39 (18): e103922. doi:10.15252/embj.2019103922. PMC 7507497. PMID 32812257.
- ^ "Watch me unveil "synthetic life"". 21 May 2010.
