Peptide synthesis: Difference between revisions
Script-assisted style fixes: mainly date formats |
person's name |
||
| (One intermediate revision by one other user not shown) | |||
| Line 2: | Line 2: | ||
In [[organic chemistry]], '''peptide synthesis''' is the production of [[peptide]]s, which are [[organic compound]]s in which multiple [[amino acid]]s are linked via amide bonds which are also known as [[peptide bond]]s. The biological process of producing long peptides (proteins) is known as [[protein biosynthesis]]. |
In [[organic chemistry]], '''peptide synthesis''' is the production of [[peptide]]s, which are [[organic compound]]s in which multiple [[amino acid]]s are linked via amide bonds which are also known as [[peptide bond]]s. The biological process of producing long peptides (proteins) is known as [[protein biosynthesis]]. |
||
[[Image:Peptide Synthesis.svg|thumb|right|500px|upright=4.1|alt=Table of Amino Acids.| Solid- |
[[Image:Peptide Synthesis.svg|thumb|right|500px|upright=4.1|alt=Table of Amino Acids.| Solid-phase peptide synthesis on a Rink amide [[resin]] using [[Fmoc]]-α-[[amine]]-protected [[amino acid]]]] |
||
== Chemistry == |
== Chemistry == |
||
Revision as of 09:49, 2 June 2013
In organic chemistry, peptide synthesis is the production of peptides, which are organic compounds in which multiple amino acids are linked via amide bonds which are also known as peptide bonds. The biological process of producing long peptides (proteins) is known as protein biosynthesis.

Chemistry
Peptides are synthesized by coupling the carboxyl group or C-terminus of one amino acid to the amino group or N-terminus of another. Due to the possibility of unintended reactions, protecting groups are usually necessary. Chemical peptide synthesis starts at the C-terminal end of the peptide and ends at the N-terminus. This is the opposite of protein biosynthesis, which starts at the N-terminal end.
Liquid-phase synthesis
Liquid-phase peptide synthesis is a classical approach to peptide synthesis. It has been replaced in most labs by solid-phase synthesis (see below). However, it retains usefulness in large-scale production of peptides for industrial purposes.
Solid-phase synthesis
Solid-phase peptide synthesis (SPPS), pioneered by Robert Bruce Merrifield,[1] resulted in a paradigm shift within the peptide synthesis community. It is now the accepted method for creating peptides and proteins in the lab in a synthetic manner. SPPS allows the synthesis of natural peptides which are difficult to express in bacteria, the incorporation of unnatural amino acids, peptide/protein backbone modification, and the synthesis of D-proteins, which consist of D-amino acids.
Small solid beads, insoluble yet porous, are treated with functional units ('linkers') on which peptide chains can be built. The peptide will remain covalently attached to the bead until cleaved from it by a reagent such as anhydrous hydrogen fluoride or trifluoroacetic acid. The peptide is thus 'immobilized' on the solid-phase and can be retained during a filtration process, whereas liquid-phase reagents and by-products of synthesis are flushed away.
The general principle of SPPS is one of repeated cycles of coupling-wash-deprotection-wash. The free N-terminal amine of a solid-phase attached peptide is coupled (see below) to a single N-protected amino acid unit. This unit is then deprotected, revealing a new N-terminal amine to which a further amino acid may be attached. The superiority of this technique partially lies in the ability to perform wash cycles after each reaction, removing excess reagent with all of the growing peptide of interest remaining covalently attached to the insoluble resin.
The overwhelmingly important consideration is to generate extremely high yield in each step. For example, if each coupling step were to have 99% yield, a 26-amino acid peptide would be synthesized in 77% final yield (assuming 100% yield in each deprotection); if each step were 95%, it would be synthesized in 25% yield. Thus each amino acid is added in major excess (2~10x) and coupling amino acids together is highly optimized by a series of well-characterized agents.
There are two majorly used forms of SPPS – Fmoc and Boc. Unlike ribosome protein synthesis, solid-phase peptide synthesis proceeds in a C-terminal to N-terminal fashion. The N-termini of amino acid monomers is protected by either of these two groups and added onto a deprotected amino acid chain.
Automated synthesizers are available for both techniques, though many research groups continue to perform SPPS manually.
SPPS is limited by yields, and typically peptides and proteins in the range of 70 amino acids are pushing the limits of synthetic accessibility. Synthetic difficulty also is sequence dependent; typically amyloid peptides and proteins are difficult to make. Longer lengths can be accessed by using native chemical ligation to couple two peptides together with quantitative yields.
Since its introduction over 40 years ago, SPPS has been significantly optimized. First, the resins themselves have been optimized.[2] Furthermore, the 'linkers' between the C-terminal amino acid and polystyrene resin have improved attachment and cleavage to the point of mostly quantitative yields.[3][4][5] The evolution of side chain protecting groups has limited the frequency of unwanted side reactions. In addition, the evolution of new activating groups on the carboxyl group of the incoming amino acid have improved coupling and decreased epimerization. Finally, the process itself has been optimized. In Merrifield's initial report, the deprotection of the α-amino group resulted in the formation of a peptide-resin salt, which required neutralization with base prior to coupling. The time between neutralization of the amino group and coupling of the next amino acid allowed for aggregation of peptides, primarily through the formation of secondary structures, and adversely affected coupling. The Kent group showed that concomitant neutralization of the α-amino group and coupling of the next amino acid led to improved coupling.[6] Each of these improvements has helped SPPS become the robust technique that it is today.
BOP SPPS
The use of BOP reagent was first described by Castro et al. in 1975.
Solid supports
The name solid support implies that reactions are carried out on the surface of the support, but this is not the case. Reactions also occur within these particles, and thus the term "solid support" better describes the insolubility of the polymer. The physical properties of the solid support, and the applications to which it can be utilized, vary with the material from which the support is constructed, the amount of cross-linking, as well as the linker and handle being used. Most scientists in the field believe that supports should have the minimum amount of cross-linking to confer stability. This should result in a well-solvated system where solid-phase peptide synthesis can be carried out. Nonetheless, the characteristics of an efficient solid support include:[7]
- It must be physically stable and permit the rapid filtration of liquids, such as excess reagents
- It must be inert to all reagents and solvents used during SPPS
- It must swell extensively in the solvents used to allow for penetration of the reagents
- It must allow for the attachment of the first amino acid
There are four primary types of solid supports:[7]
- Gel-type supports: These are highly solvated polymers with an equal distribution of functional groups. This type of support is the most common, and includes:
- Polystyrene: Styrene cross-linked with 1–2% divinylbenzene
- Polyacrylamide: A hydrophilic alternative to polystyrene
- Polyethylene glycol (PEG): PEG-Polystyrene (PEG-PS) is more stable than polystyrene and spaces the site of synthesis from the polymer backbone
- PEG-based supports: Composed of a PEG-polypropylene glycol network or PEG with polyamide or polystyrene
- Surface-type supports: Many materials have been developed for surface functionalization, including controlled pore glass, cellulose fibers, and highly cross-linked polystyrene.
- Composites: Gel-type polymers supported by rigid matrices.
Polystyrene resin
Polystyrene resin is a versatile resin and it is quite useful in multi-well, automated peptide synthesis, due to its minimal swelling in dichloromethane. The initial support used by R. Bruce Merrifield was polysytrene cross-linked with 2% divinylbenzene. This support is sometimes referred to as the 'Merrifield resin.' This produces a hydrophobic bead that is solvated by nonpolar solvent such as dichloromethane. Since then, new resins have been developed with the following advantages:
- Enhanced swelling or rigidity (a property of mechanical strength)
- Chemical inertness
Highly cross-linked (50%) polystyrene has been developed that possesses the features of increased mechanical stability, better filtration of reagents and solvents, and rapid reaction kinetics.
Polyamide resin
Polyamide resin is also a useful and versatile resin. It seems to swell much more than polystyrene, in which case it may not be suitable for some automated synthesizers, if the wells are too small.
PEG hybrid polystyrene resin
An example of this type of resin is the Tentagel resin. The base resin is polystyrene onto which is attached long chains (Mw ca. 3000 Da) of polyethylene glycol (PEG; also known as polyethylene oxide). Synthesis is carried out on the distal end of the PEG spacer making it suited for long and difficult peptides. In addition it is also attractive for the synthesis of combinatorial Peptide libraries and on resin screening experiments. It does not expand much during synthesis making it a preferred resin for robotic peptide synthesis.
PEG-based resin
ChemMatrix(R) is a new type of resin which is based on PEG that is crosslinked. ChemMatrix(R) has claimed a high chemical and thermal stability (is compatible with Microwave synthesis) and has shown higher degrees of swellings in acetonitrile, dichloromethane, DMF, N-methylpyrrolidone, TFA and water compared to the polystyrene based resins. ChemMatrix has shown significant improvements to the synthesis of hydrophobic sequences. ChemMatrix may be useful for the synthesis of difficult and long peptides.
Improvements to solid supports used for peptide synthesis enhance their ability to withstand the repeated use of TFA during the deprotection step of SPPS.[8] Furthermore, different resins allow for different functional groups at the C-terminus. The oxymethylphenylacetamidomethyl (PAM) resin results in the conventional C-terminal carboxylic acid. On the other hand, the paramethylbenzhydrylamine (pMBHA) resin yields a C-terminal amide, which is useful in mimicking the interior of a protein.
Along with the development of Fmoc SPPS, different resins have also been created to be removed by TFA. Similar to the Boc strategy, two primary resins are used, based on whether a C-terminal carboxylic acid or amide is desired. The Wang resin is the most commonly used resin for peptides with C-terminal carboxylic acids.[9] If a C-terminal amide is desired, the Rink amide resin is used.
Protecting groups
Amino acids have reactive moieties at the N- and C-termini, which facilitates amino acid coupling during synthesis. Many amino acids also have reactive side chain functional groups, which can interact with free termini or other side chain groups during synthesis and peptide elongation and negatively influence yield and purity. To facilitate proper amino acid synthesis with minimal side chain reactivity, chemical groups have been developed to bind to specific amino acid functional groups and block, or protect, the functional group from nonspecific reactions. These protecting groups, while vast in nature, can be separated into three groups, as follows:
- N-terminal protecting groups
- C-terminal protecting groups (mostly used in liquid-phase synthesis)
- Side chain protecting groups
Purified, individual amino acids are reacted with these protecting groups prior to synthesis and then selectively removed during specific steps of peptide synthesis.
N-terminal protecting groups
Amino acids are added to excess to ensure complete coupling during each synthesis step, and without N-terminal protection, polymerization of unprotected amino acids could occur, resulting in low peptide yield or synthesis failure. N-terminal protection requires an additional step of removing the protecting group, termed deprotection, prior to the coupling step, creating a repeating design flow as follows:
- Protecting group is removed from the trailing amino acids in a deprotection reaction
- Deprotection reagents are washed away to provide a clean coupling environment
- Protected amino acids dissolved in a solvent such as dimethylformamide (DMF) combined with coupling reagents are pumped through the synthesis column
- Coupling reagents are washed away to provide clean deprotection environment
Currently, two protecting groups (t-Boc, Fmoc) are commonly used in solid-phase peptide synthesis. Their lability is caused by the carbamate group which readily releases CO2 for an irreversible decoupling step.
t-Boc protecting group

The original method for the synthesis of proteins relied on (tert-butyloxycarbonyl or more simply "Boc") to temporarily protect the α-amino group. In this method, the Boc group is covalently bound to the amino group to suppress its nucleophilicity. The C-terminal amino acid is covalently linked to the resin through a linker. Next, the Boc group is removed with acid, such as trifluoroacetic acid (TFA). This forms a positively-charged amino group (in the presence of excess TFA; note image on the right illustrates neutral amino group), which is neutralized (via in-situ or non-in-situ methods) and coupled to the incoming activated amino acid.[6] Reactions are driven to completion by the use of excess (two- to four-fold) activated amino acid. After each deprotection and coupling step, a wash with N,N-dimethylformamide (DMF) is performed to remove excess reagents, allowing for high yields (~99%) during each cycle.[7]
t-Boc protecting strategies retain usefulness in reducing peptide aggregation during synthesis. t-Boc groups can be added to amino acids with t-Boc anhydride and a suitable base. Some researchers prefer Boc SPPS for complex syntheses . In addition, when synthesizing nonnatural peptide analogs, which are base-sensitive (such as depsipeptides), the t-Boc protecting group is necessary, because Fmoc SPPS uses a base to deprotect the α-amino group.
Permanent side-chain protecting groups are typically benzyl or benzyl-based groups. Final removal of the peptide from the linkage occurs simultaneously with side-chain deprotection with anhydrous hydrogen fluoride via hydrolytic cleavage. The final product is a fluoride salt which is relatively easy to solubilize. Importantly, scavengers such as cresol are added to the HF in order to prevent reactive t-butyl cations from generating undesired products. In fact, the use of harsh hydrogen fluoride may degrade some peptides, which was the premise for the development of a milder, base-labile method of SPPS—namely, the Fmoc method.
Fmoc protecting group

The capacity for anhydrous hydrogen fluoride to degrade proteins during the final cleavage conditions led to a new α-amino protecting group based on 9-fluorenylmethyloxycarbonyl (Fmoc). The Fmoc method allows for a milder deprotection scheme. This method utilizes a base, usually piperidine (20–50%) in DMF in order to remove the Fmoc group to expose the α-amino group for reaction with an incoming activated amino acid.[7] Unlike the acid used to deprotect the α-amino group in Boc methods, Fmoc SPPS uses a base, and thus the exposed amine is neutral. Therefore, no neutralization of the peptide-resin is required, but the lack of electrostatic repulsions between the peptides can lead to increased aggregation. Because the liberated fluorenyl group is a chromophore, deprotection by Fmoc can be monitored by UV absorbance of the runoff, a strategy which is employed in automated synthesizers.
The advantage of Fmoc is that it is cleaved under very mild basic conditions (e.g. piperidine), but stable under acidic conditions, although this has not always held true in certain synthetic sequences. This allows mild acid-labile protecting groups that are stable under basic conditions, such as Boc and benzyl groups, to be used on the side-chains of amino acid residues of the target peptide. This orthogonal protecting group strategy is common in organic synthesis. Fmoc is preferred over BOC due to ease of cleavage; however it is less atom-economical, as the fluorenyl group is much larger than the tert-butyl group. Accordingly, prices for Fmoc amino acids were high until the large-scale piloting of one of the first synthesized peptide drugs, enfuvirtide, began in the 1990s, when market demand adjusted the relative prices of the two sets of amino acids.
Semipermanent side chain protecting groups are t-butyl based, and final cleavage of the protein from the resin and removal of permanent protecting groups is performed with TFA in the presence of scavengers. Water and triisopropylsilane (TIPS) present in a 1:1 ratio are often used as scavengers. Thus, the Fmoc method is orthogonal in two directions: deprotection of any α-amino group, deprotection of side groups and final cleavage from the resin occur by independent mechanisms. The resulting final product is a TFA salt, which is more difficult to solubilize than the fluoride salts generated in Boc SPPS. This method is thus milder than the Boc method because the deprotection/cleavage-from-resin steps occur with different conditions rather than with different reaction rates.
Comparison of Boc and Fmoc solid-phase peptide synthesis
Both the Fmoc and Boc methods offer advantages and disadvantages. The selection of one technique over another is thus made on a case-by-case basis.[10]
| Boc | Fmoc | |
|---|---|---|
| Requires special equipment | Yes | No |
| Cost of reagents | Lower | Higher |
| Solubility of peptides | Higher | Lower |
| Purity of hydrophobic peptides | High | May be lower |
| Problems with aggregation | Less frequently | More frequently |
| Synthesis time | ~20 min/amino acid | ~20–60 min/amino acid |
| Final deprotection | HF | TFA |
| Safety | Potentially dangerous | Relatively safe |
| Orthogonal | No | Yes |
Boc SPPS uses special equipment to handle the final cleavage and deprotection step, which requires anhydrous hydrogen fluoride. Because the final cleavage of the peptide with Fmoc SPPS uses TFA, this special equipment is not necessary. The solubility of peptides generated by Boc SPPS is generally higher than those generated with the Fmoc method, because fluoride salts are higher in solubility than TFA salts. Next, problems with aggregation are generally more of an issue with Fmoc SPPS. This is primarily because the removal of a Boc group with TFA yields a positively-charged α-amino group, whereas the removal of an Fmoc group yields a neutral α-amino group. The steric hindrance of the positively charged α-amino group limits the formation of secondary structure on the resin. Finally, the Fmoc method is considered orthogonal, since α-amino group deprotection is with base, while final cleavage from the resin is with acid. The Boc method utilizes acid for both deprotection and cleavage from the resin. Based on this comparison, one sees that both methods possess advantages and disadvantages. Thus, several factors help to decide which method may be preferable.
DMF must be 'peptide grade' i.e. little/no impurities and must also be 'fresh'. This is due to the fact that DMF undergoes photolysis to form carbon monoxide and dimethylamine. Dimethylamine may remove the Fmoc group and, therefore, lead to impurities.
Benzyloxy-carbonyl (Z) group
The first use of (Z) group as protecting groups was done by Max Bergmann who synthesised oligopeptides.[11]
Another carbamate based group is the benzyloxy-carbonyl (Z) group. It is removed in harsher conditions: HBr/acetic acid or catalytic hydrogenation. Today it is almost exclusively used for side chain protection.
Alloc protecting group
The allyloxycarbonyl (alloc) protecting group is often used to protect a carboxylic acid, hydroxyl, or amino group when an orthogonal deprotection scheme is required. It is sometimes used when conducting on-resin cyclic peptide formation, where the peptide is linked to the resin by a side-chain functional group. The alloc group can be removed using tetrakis(triphenylphosphine)palladium(0) along with a 37:2:1 mixture of methylene chloride, acetic acid, and N-Methylmorpholine (NMM) for 2 hours. The resin must then be carefully washed 0.5% DIPEA in DMF, 3 × 10 ml of 0.5% sodium diethylthiocarbamate in DMF, and then 5 × 10 ml of 1:1 DCM:DMF.
Lithographic protecting groups
For special applications like protein microarrays lithographic protecting groups are used. Those groups can be removed through exposure to light.
Side chain protecting groups
Amino acid side chains represent a broad range of functional groups and are sites of nonspecific reactivity during peptide synthesis. Because of this, many different protecting groups are required that are usually based on the benzyl (Bzl) or tert-butyl (tBu) group.[7] The specific protecting groups used during the synthesis of a given peptide vary depending on the peptide sequence and the type of N-terminal protection used (see next paragraph). Side chain protecting groups are known as permanent or semipermanent protecting groups, because they can withstand the multiple cycles of chemical treatment during synthesis and are only removed during treatment with strong acids after peptide synthesis is completed.
Protection schemes
Because multiple protecting groups are normally used during peptide synthesis, these groups must be compatible to allow deprotection of distinct protecting groups while not affecting other protecting groups. Protecting schemes are therefore established to match protecting groups so that deprotection of one protecting group does not affect the binding of the other groups. Because N-terminal deprotection occurs continuously during peptide synthesis, protecting schemes have been established in which the different types of side chain protecting groups (Bzl or tBu) are matched to either Boc or Fmoc, respectively, for optimized deprotection. These protecting schemes also incorporate each of the steps of synthesis and cleavage, as described in the table and in later sections of this page.
Activating groups
For coupling the peptides the carboxyl group is usually activated. This is important for speeding up the reaction. There are two main types of activating groups: carbodiimides and triazolols. However the use of pentafluorophenyl esters (FDPP,[12] PFPOH[13]) and BOP-Cl[14] are useful for cyclising peptides.
Carbodiimides

These activating agents were first developed. Most common are dicyclohexylcarbodiimide (DCC) and diisopropylcarbodiimide (DIC). Reaction with a carboxylic acid yields a highly reactive O-acylisourea. During artificial protein synthesis (such as Fmoc solid-state synthesizers), the C-terminus is often used as the attachment site on which the amino acid monomers are added. To enhance the electrophilicity of carboxylate group, the negatively charged oxygen must first be "activated" into a better leaving group. DCC is used for this purpose. The negatively charged oxygen will act as a nucleophile, attacking the central carbon in DCC. DCC is temporarily attached to the former carboxylate group (which is now an ester group), making nucleophilic attack by an amino group (on the attaching amino acid) to the former C-terminus (carbonyl group) more efficient. The problem with carbodiimides is that they are too reactive and that they can therefore cause racemization of the amino acid.
Triazoles
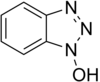
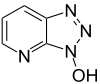
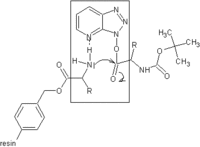
To solve the problem of racemization, triazoles were introduced. The most important ones are 1-hydroxy-benzotriazole (HOBt) and 1-hydroxy-7-aza-benzotriazole (HOAt). Others have been developed. These substances can react with the O-acylurea to form an active ester which is less reactive and less in danger of racemization. HOAt is especially favourable because of a neighbouring group effect.[15] Recently, HOBt has been removed from many chemical vendor catalogues; although almost always found as a hydrate, HOBt may be explosive when allowed to fully dehydrate and shipment by air or sea is heavily restricted. Alternatives to HOBt and HOAt have been introduced. One of the most promising and inexpensive [citation needed] is ethyl 2-cyano-2-(hydroxyimino)acetate (trade name Oxyma Pure), which is not explosive and has a reactivity of that in between HOBt and HOAt.

Newer developments omit the carbodiimides totally. The active ester is introduced as a uronium or phosphonium salt of a non-nucleophilic anion (tetrafluoroborate or hexafluorophosphate): HBTU, HATU, HCTU, TBTU, PyBOP. Two uronium types of the coupling additive of Oxyma Pure is also available as COMU or TOTU reagent.
Regioselective disulfide formation
The formation of multiple native disulfides remains one of the primary challenges of native peptide synthesis by solid-phase methods. Random chain combination typically results in several products with nonnative disulfide bonds.[16] Stepwise formation of disulfide bonds is typically the preferred method, and performed with thiol protecting groups (PGs).[17] Different thiol PGs provide multiple dimensions of orthogonal protection. These orthogonally-protected cysteines are incorporated during the solid-phase synthesis of the peptide. Successive removal of these PGs to allow for selective exposure of free thiol groups, leads to disulfide formation in a stepwise manner. The order of removal of these PGs must be considered so that only one group is removed at a time. Using this method, Kiso et al. reported the first total synthesis of insulin by this method in 1993.[18]
The thiol PGs must possess multiple characteristics. First, the PG must be reversible with conditions that do not affect the unprotected side chains. Second, the protecting group must be able to withstand the conditions of solid-phase synthesis. Third, the configuration of the removal of the thiol protecting group must be such that it leaves intact other thiol PGs, if orthogonal protection is desired. That is, the removal of PG A should not affect PG B. Some of the thiol PGs commonly used include the acetamidomethyl (Acm), tert-butyl (But), 3-nitro-2-pyridine sulfenyl (NPYS), 2-pyridine-sulfenyl (Pyr), and triphenylmethyl (Trt) groups. Importantly, the NPYS group can replace the Acm PG to yield an activated thiol.[19]
In the stepwise formation of disulfides to synthesize insulin by Kiso et al., the authors synthesize the A-chain with following protection: CysA6(But); CysA7(Acm); CysA11(But). Thus, CysA20 is unprotected. Synthesis of the B-chain is performed with the following protection: CysB7(Acm) CysB19(Pyr). The first disulfide bond, CysA20–CysB19, was formed by mixing the two chains in 8 M urea, pH 8 (RT) for 50 min. The second disulfide bond, CysA7–CysB7, was formed by treatment with iodine in aqueous acetic acid to remove the Acm groups. The third disulfide, the intramolecular CysA6–CysA11, was formed by the removal of the But groups by methyltrichlorosilane with diphenyl sulfoxide in TFA. Importantly, formation of the first disulfide in 8 M urea, pH 8 does not affect the other PGs, namely Acm and But groups. Likewise, formation of the second disulfide bond with iodine in aqueous acetic acid does not affect the But groups.
Important to the discussion of disulfide bond formation is the order in which disulfides are formed. From a logical standpoint, the order in which the thiol groups are exposed to form disulfides should be of little consequence, since the other cysteines are protected. Practically, however, the order in which disulfides are formed can have a significant effect on yields. This may be because the formation of the CysA20–CysB19 disulfide may place the thiol group of CysB7 in close proximity with both CysA6 and CysA7, leading to multiple disulfide products. This is one manifestation of the reality that solid-phase peptide synthesis is as much art as it is science.
Synthesizing long peptides
Stepwise elongation, in which the amino acids are connected step-by-step in turn, is ideal for small peptides containing between 2 and 100 amino acid residues. Another method is fragment condensation, in which peptide fragments are coupled. Although the former can elongate the peptide chain without racemization, the yield drops if only it is used in the creation of long or highly polar peptides. Fragment condensation is better than stepwise elongation for synthesizing sophisticated long peptides, but its use must be restricted in order to protect against racemization. Fragment condensation is also undesirable since the coupled fragment must be in gross excess, which may be a limitation depending on the length of the fragment.
A new development for producing longer peptide chains is chemical ligation: Unprotected peptide chains react chemoselectively in aqueous solution. A first kinetically controlled product rearranges to form the amide bond. The most common form of native chemical ligation uses a peptide thioester that reacts with a terminal cysteine residue.
In order to optimize synthesis of long peptides, Zealand Pharma (located in Denmark in Medicon Valley) invented a method for converting a difficult peptide sequence into an easy peptide sequence. The new technology, called SIP-technology, uses “structure-inducing probes” (SIP) to facilitate the synthesis of long peptides.[20] The SIP-technology is a small pre-sequence peptide sequence (e.g. Lysine (Lysn); Glutamic Acid (Glun); (LysGlu)n) that is incorporated at the C-terminus of subsequent resin bound peptide to induce an alpha-helix-like structure in the peptide. The SIP technology constrains the parent peptide into a more ordered conformation using intramolecular hydrogen bonds. This allows the peptide structure to stabilize, and the utilized hydrogen bonds reduce the likelihood of aggregation and degradation by enzymes. In this way, the SIP technology is designed to optimize peptide synthesis, increase biological half-life, improve peptide stability and inhibit enzymatic degradation without altering pharmacological activity or profile of action.[21][22]
Coupling efficiency vs. peptide length
Peptide
| Length | Coupling Efficiency | Coupling Efficiency | Coupling Efficiency | Coupling Efficiency | Coupling Efficiency |
| 1 | 0.995 | 0.99 | 0.98 | 0.97 | 0.96 |
| 5 | 0.98 | 0.95 | 0.92 | 0.89 | 0.85 |
| 10 | 0.96 | 0.91 | 0.83 | 0.76 | 0.69 |
| 15 | 0.93 | 0.87 | 0.75 | 0.65 | 0.56 |
| 20 | 0.91 | 0.83 | 0.68 | 0.56 | 0.46 |
| 25 | 0.89 | 0.79 | 0.62 | 0.48 | 0.38 |
| 30 | 0.86 | 0.75 | 0.56 | 0.41 | 0.31 |
| 35 | 0.84 | 0.71 | 0.50 | 0.36 | 0.25 |
| 40 | 0.82 | 0.67 | 0.45 | 0.30 | 0.20 |
| 45 | 0.80 | 0.63 | 0.41 | 0.26 | 0.17 |
| 50 | 0.78 | 0.60 | 0.37 | 0.22 | 0.14 |
| 55 | 0.76 | 0.58 | 0.34 | 0.19 | 0.11 |
| 60 | 0.74 | 0.55 | 0.30 | 0.17 | 0.09 |
| 65 | 0.73 | 0.53 | 0.27 | 0.14 | 0.07 |
| 70 | 0.71 | 0.50 | 0.25 | 0.12 | 0.06 |
Microwave assisted peptide synthesis
Although microwave irradiation has been around since the late 1940s, it was not until 1986 that microwave energy was used in organic chemistry. During the end of the 1980s and 1990s, microwave energy was an obvious source for completing chemical reactions in minutes that would otherwise take several hours to days. Through several technical improvements at the end of the 1990s and beginning of the 2000s, microwave synthesizers have been designed to provide both low and high energy pockets of microwave energy so that the temperature of the reaction mixture could be controlled. The microwave energy used in peptide synthesis is of a single frequency providing maximum penetration depth of the sample which is in contrast to conventional kitchen microwaves.
In peptide synthesis, microwave irradiation has been used to complete long peptide sequences with high degrees of yield and low degrees of racemization.[23] Microwave irradiation during the coupling of amino acids to a growing polypeptide chain is not only catalyzed through the increase in temperature, but also due to the alternating electromagnetic radiation[citation needed] to which the polar backbone of the polypeptide continuously aligns to. Due to this phenomenon, the microwave energy can prevent aggregation and thus increases yields of the final peptide product. There is however no clear evidence that microwave is better than simple heating and some peptide laboratories regard microwave just as a convenient method for rapid heating of the peptidyl resin. Heating to above 50–55 degrees Celsius also prevents aggregation and accelerates the coupling.
Despite the main advantages of microwave irradiation of peptide synthesis, the main disadvantage is the racemization which may occur with the coupling of cysteine and histidine. A typical coupling reaction with these amino acids are performed at lower temperatures than the other 18 natural amino acids. A number of peptides do not survive microwave synthesis or heating in general. One of the more serious side effects is dehydration (loss of water) which for certain peptides can be almost quantitative like pancreatic polypeptide (PP). This side effect is also seen by simple heating without the use of microwave.
Cyclic peptides
On resin cyclization
Peptide can be cyclize on solid support. A variety of cylization reagents can be used such as HBTU/HOBt/DIEA, PyBop/DIEA, PyClock/DIEA. Head-to-tail peptides can be made on the solid support. The deprotection of the C-terminus at some suitable point allows on-resin cyclization by amide bond formation with the deprotected N-terminus. Once cyclization has taken place, the peptide is cleaved from resin by acidolysis and purified. The strategy for the solid-phase synthesis of cyclic peptides in not limited to attachment through Asp, Glu or Lys side chains. Cysteine has a very reactive sulfhydryl group on its side chain. A disulfide bridge is created when a sulfur atom from one Cysteine forms a single covalent bond with another sulfur atom from a second cysteine in a different part of the protein. These bridges help to stabilize proteins, especially those secreted from cells. Some researchers use modified cysteines using S-acetomidomethyl (Acm) to block the formation of the disulfide bond but preserve the cysteine and the protein's original primary structure.
This section is empty. You can help by adding to it. (January 2011) |
Solution phase cyclization
This section is empty. You can help by adding to it. (January 2011) |
See also
References
- ^ R. B. Merrifield (1963). "Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide". J. Am. Chem. Soc. 85 (14): 2149–2154. doi:10.1021/ja00897a025.
- ^ Mitchell, A. R. K., S.B.H.; Engelhard, M.; Merrifield, R.B. (1978). "A new synthetic route to tert-butyloxycarbonylaminoacyl-4-(oxymethyl)phenylacetamidomethyl-resin, an improved support for solid-phase peptide synthesis". J. Org. Chem. 43 (13): 2845–2852. doi:10.1021/jo00408a022.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang, S.-S. (1973). "p-alkoxybenzyl alcohol resin and p-alkoxybenzyloxycarbonylhydrazide resin for solid phase synthesis of protected peptide fragments". J. Am. Chem. Soc. 95 (4): 1328–33. PMID 4687686.
- ^ Matsueda, G. R. a. S., J.M. (1981). "A p-methylbenzylhydrlamine resin for improved solid-phase synthesis of peptide amides". Peptides. 2 (1): 45–50. doi:10.1016/S0196-9781(81)80010-1. PMID 7243625.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sieber, P. (1987). "A new acid-labile anchor group for the solid-phase synthesis of C-terminal peptide amides by the Fmoc method". Tetrahedron Lett. 34: 1269–70.
- ^ a b Schnolzer, M. A., P.; Jones, A.; Alewood, D.; Kent, S.B.H. (2007). "In Situ Neutralization in Boc-chemistry Solid Phase Peptide Synthesis". Int. J. Peptide Res. Therap. 13 (1–2): 31–44. doi:10.1007/s10989-006-9059-7.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e Albericio, F. (2000). Solid-Phase Synthesis: A Practical Guide (1 ed.). Boca Raton: CRC Press. p. 848. ISBN 0-8247-0359-6.
- ^ Feinberg, R. S.; Merrifield, R. B. (1974). "Zinc chloride-catalyzed chloromethylation of resins for solid phase peptide synthesis". Tetrahedron (17): 3209–3212. doi:10.1016/S0040-4020(01)97575-1.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hermkens, P. H. H.; Ottenheijm, H. C. J.; Rees, D. C. (1997). "Solid-phase organic reactions II: A review of the literature Nov 95 – Nov 96". Tetrahedron. 53 (16): 5643–5678. doi:10.1016/S0040-4020(97)00279-2.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nilsson BL, Soellner MB, Raines RT (2005). "Chemical Synthesis of Proteins". Annu. Rev. Biophys. Biomol. Struct. 34: 91–118. doi:10.1146/annurev.biophys.34.040204.144700. PMC 2845543. PMID 15869385.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Max Bergmann, Leonidas Zervas (1932). "Über ein allgemeines Verfahren der Peptid-Synthese". Berichte der deutschen chemischen Gesellschaft. 65 (7): 1192–1201. doi:10.1002/cber.19320650722.
- ^ K. C. Nicolaou; Natarajan, Swaminathan; Li, Hui; Jain, Nareshkumar F.; Hughes, Robert; Solomon, Michael E.; Ramanjulu, Joshi M.; Boddy, Christopher N. C.; Takayanagi, Masaru (1998). "Total Synthesis of Vancomycin Aglycon – Part 1: Synthesis of Amino Acids 4–7 and Construction of the AB-COD Ring Skeleton". Angew. Chem. Int. Ed. 37 (19): 2708–2714. doi:10.1002/(SICI)1521-3773(19981016)37:19<2708::AID-ANIE2708>3.0.CO;2-E.
- ^ Schmidt (1998). "Synthetic studies of 14-membered cyclopeptide alkaloids". Tetrahedron Lett. 39 (40): 7211–7214. doi:10.1016/S0040-4039(98)01589-5.
- ^ R. Baker, J. L. Castro (1989). "The total synthesis of (+)-macbecin I". Chem. Commun. (6): 378–381. doi:10.1039/C39890000378.
- ^ L. A. Carpino (1993). "1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive". J. Am. Chem. Soc. 115 (10): 4397–4398. doi:10.1021/ja00063a082.
- ^ Zhang, J.-W.; Wu, Cui Rong; Liu, Wen; Zhang, Jing Wen (1991). "Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications". J. Am. Chem. Soc. 113 (17): 6657–6662. doi:10.1021/ja00017a044.
- ^ Sieber, P.; Kamber, B.; Hartmann, A.; Jöhl, A.; Riniker, B.; Rittel, W. (1977). "Total synthesis of human insulin. IV. Description of the final steps (author's transl)". Helvetica Chimica Acta. 60 (1): 27–37. doi:10.1002/hlca.19770600105. PMID 838597.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Akaji, K.; Fujino, K.; Tatsumi, T.; Kiso, Y. (1993). "Total synthesis of human insulin by regioselective disulfide formation using the silyl chloride-sulfoxide method". Journal of the American Chemical Society. 115 (24): 11384–11392. doi:10.1021/ja00077a043.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ottl, J.; Battistuta, R.; Pieper, M.; Tschesche, H.; Bode, W.; Kuhn, K.; Moroder, L. (1996). "Design and synthesis of heterotrimeric collagen peptides with a built-in cystine-knot. Models for collagen catabolism by matrix-metalloproteases". FEBS Lett. 398 (1): 31–36. doi:10.1016/S0014-5793(96)01212-4. PMID 8946948.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ http://www.zealandpharma.com/%7B2557DE5E-28D5-4782-A161-B8EC9836CB43%7D
- ^ Kapusta, D. R.; Thorkildsen, C; Kenigs, VA; Meier, E; Vinge, MM; Quist, C; Petersen, JS (2005). "Pharmacodynamic Characterization of ZP120 (Ac-RYYRWKKKKKKK-NH2), a Novel, Functionally Selective Nociceptin/Orphanin FQ Peptide Receptor Partial Agonist with Sodium-Potassium-Sparing Aquaretic Activity". Journal of Pharmacology and Experimental Therapeutics. 314 (2): 652–60. doi:10.1124/jpet.105.083436. PMID 15855355.
- ^ Rizzi A, Rizzi D, Marzola G; et al. (2002). "Pharmacological characterization of the novel nociceptin/orphanin FQ receptor ligand, ZP120: in vitro and in vivo studies in mice". Br. J. Pharmacol. 137 (3): 369–74. doi:10.1038/sj.bjp.0704894. PMC 1573505. PMID 12237257.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Stacey A. Palasek, Zachary J. Cox, Jonathan M. Collins (2007). "Limiting racemization and aspartimide formation in microwave-enhanced Fmoc solid phase peptide synthesis". Journal of Peptide Science. 13 (3): 143–148. doi:10.1002/psc.804. PMID 17121420.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- Atherton, E.; Sheppard, R.C. (1989). Solid Phase peptide synthesis: a practical approach. Oxford, England: IRL Press. ISBN 0-19-963067-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - Stewart, J.M.; Young, J.D. (1984). Solid phase peptide synthesis (2nd ed.). Rockford: Pierce Chemical Company. p. 91. ISBN 0-935940-03-0.
{{cite book}}: CS1 maint: multiple names: authors list (link)
External links
- Peptide Calculator from Bio-Synthesis,biosyn.com
- Amide coupling – Synthetic protocols from organic-reaction.com
- Further information concerning peptide synthesis from AltaBioscience
For introduction of protecting group and mechanism of deprotection See : http://www.biocis.u-psud.fr/spip.php?article332
