AlphaFold: Difference between revisions
mNo edit summary |
→Responses: this too; please use WP:MEDRS sources for extraordinary claims |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
In November 2020, a version of the program titled AlphaFold 2 took part in the 14th edition of the biennial [[Critical Assessment of Techniques for Protein Structure Prediction]] ([[CASP]]) competition,<ref>{{Cite web|last=Shead|first=Sam|date=2020-11-30|title=DeepMind solves 50-year-old 'grand challenge' with protein folding A.I.|url=https://www.cnbc.com/2020/11/30/deepmind-solves-protein-folding-grand-challenge-with-alphafold-ai.html|access-date=2020-11-30|website=CNBC|language=en}}</ref> in which it achieved a level of accuracy much higher than any other computational method.<ref name=":0" /> The program scored above 90 for around two-thirds of the proteins in CASP's [[Global distance test|global distance test (GDT)]], a test that measures the degree to which a computational program predicted structure is similar to the lab experiment determined structure, with 100 being an exact match, within the distance cutoff used for calculating GDT.<ref name=":0" /><ref name=":4">Robert F. Service, [https://www.sciencemag.org/news/2020/11/game-has-changed-ai-triumphs-solving-protein-structures ‘The game has changed.’ AI triumphs at solving protein structures], ''[[Science (magazine)|Science]]'', 30 November 2020</ref> |
In November 2020, a version of the program titled AlphaFold 2 took part in the 14th edition of the biennial [[Critical Assessment of Techniques for Protein Structure Prediction]] ([[CASP]]) competition,<ref>{{Cite web|last=Shead|first=Sam|date=2020-11-30|title=DeepMind solves 50-year-old 'grand challenge' with protein folding A.I.|url=https://www.cnbc.com/2020/11/30/deepmind-solves-protein-folding-grand-challenge-with-alphafold-ai.html|access-date=2020-11-30|website=CNBC|language=en}}</ref> in which it achieved a level of accuracy much higher than any other computational method.<ref name=":0" /> The program scored above 90 for around two-thirds of the proteins in CASP's [[Global distance test|global distance test (GDT)]], a test that measures the degree to which a computational program predicted structure is similar to the lab experiment determined structure, with 100 being an exact match, within the distance cutoff used for calculating GDT.<ref name=":0" /><ref name=":4">Robert F. Service, [https://www.sciencemag.org/news/2020/11/game-has-changed-ai-triumphs-solving-protein-structures ‘The game has changed.’ AI triumphs at solving protein structures], ''[[Science (magazine)|Science]]'', 30 November 2020</ref> |
||
AlphaFold 2's results at CASP were described as "astounding",<ref name=AlQuraishiTweet /> transformational |
AlphaFold 2's results at CASP were described as "astounding",<ref name=AlQuraishiTweet /> and transformational.<ref name=":5" /> Others have objected that any such statement is premature, given that AlphaFold's method has not yet been made known in sufficient detail to allow discussion, nor full assessment of its limitations, nor independent reimplementation; nor are the reasons for its success yet understood qualitiatively; and that considerable challenges remain before the questions of [[protein folding]] can be said to be fully understood. Nevertheless there has been widespread respect for the technical achievement. |
||
Others have objected that any such statement is premature, given that AlphaFold's method has not yet been made known in sufficient detail to allow discussion, nor full assessment of its limitations, nor independent reimplementation; nor are the reasons for its success yet understood qualitiatively; and that considerable challenges remain before the questions of [[protein folding]] can be said to be fully understood. Nevertheless there has been widespread respect for the technical achievement. |
|||
== Protein folding problem == |
== Protein folding problem == |
||
| Line 52: | Line 50: | ||
== Responses == |
== Responses == |
||
AlphaFold 2 scoring more than 90 in [[CASP]]'s [[Global distance test|global distance test (GDT)]] is considered a significant achievement in [[computational biology]].<ref name=":4" /> [[Nobel Prize in Chemistry|Nobel Prize]] winner and [[Structural biology|structural biologist]] [[Venki Ramakrishnan]] called the result "a stunning advance on the protein folding problem",<ref name=":4" /> adding that "It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research."<ref name=DeepMindAlpha2 /> |
AlphaFold 2 scoring more than 90 in [[CASP]]'s [[Global distance test|global distance test (GDT)]] is considered a significant achievement in [[computational biology]].<ref name=":4" /> [[Nobel Prize in Chemistry|Nobel Prize]] winner and [[Structural biology|structural biologist]] [[Venki Ramakrishnan]] called the result "a stunning advance on the protein folding problem",<ref name=":4" /> adding that "It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research."<ref name=DeepMindAlpha2 /> |
||
Propelled by press releases from CASP and DeepMind,<ref name=CASP_release>[https://predictioncenter.org/casp14/doc/CASP14_press_release.html Artificial intelligence solution to a 50-year-old science challenge could ‘revolutionise’ medical research] (press release), [[CASP]] organising committee, 30 November 2020</ref><ref name=DeepMindAlpha2 /> AlphaFold 2's success received wide media attention.<ref>Brigitte Nerlich, [https://blogs.nottingham.ac.uk/makingsciencepublic/2020/12/04/protein-folding-and-science-communication-between-hype-and-humility/ Protein folding and science communication: Between hype and humility], [[University of Nottingham]] blog, 4 December 2020</ref> As well as news pieces in the specialist science press, such as ''[[Nature]]'',<ref name=":5" /> ''[[Science]]'',<ref name=":4" /> ''[[MIT Technology Review]]'',<ref name=":0" /> and ''[[New Scientist]]'',<ref>Michael Le Page, [https://www.newscientist.com/article/2261156-deepminds-ai-biologist-can-decipher-secrets-of-the-machinery-of-life/ DeepMind's AI biologist can decipher secrets of the machinery of life], ''[[New Scientist]]'', 30 November 2020</ref><ref>[https://www.newscientist.com/article/2261613-the-predictions-of-deepminds-latest-ai-could-revolutionise-medicine/ The predictions of DeepMind’s latest AI could revolutionise medicine], ''[[New Scientist]]'', 2 December 2020</ref> the story was widely covered by major national newspapers,<ref>[[Cade Metz]], [https://www.nytimes.com/2020/11/30/technology/deepmind-ai-protein-folding.html London A.I. Lab Claims Breakthrough That Could Accelerate Drug Discovery], ''[[New York Times]]'', 30 November 2020</ref><ref>Ian Sample,[https://www.theguardian.com/technology/2020/nov/30/deepmind-ai-cracks-50-year-old-problem-of-biology-research DeepMind AI cracks 50-year-old problem of protein folding], ''[[The Guardian]]'', 30 November 2020</ref><ref>Lizzie Roberts, [https://www.telegraph.co.uk/news/2020/11/30/google-ai-researchers-crack-50-year-old-protein-folding-problem/ 'Once in a generation advance' as Google AI researchers crack 50-year-old biological challenge]. ''[[Daily Telegraph]]'', 30 November 2020</ref><ref name=ElPais /> as well as general news-services and weekly publications, such as ''[[Fortune (magazine)|Fortune]]'',<ref>Jeremy Kahn, [https://fortune.com/2020/11/30/deepmind-protein-folding-breakthrough/ In a major scientific breakthrough, A.I. predicts the exact shape of proteins], ''[[Fortune (magazine)|Fortune]]'', 30 November 2020</ref><ref name=KahnLessons /> ''[[The Economist]]'',<ref name=":1" /> [[Bloomberg LP|Bloomberg]],<ref name=":2" /> ''[[Der Spiegel]]'',<ref name=Spiegel_1>Julia Merlot, [https://www.spiegel.de/wissenschaft/medizin/kuenstliche-intelligenz-sagt-faltung-von-proteinen-praezise-voraus-a-c52705ef-d3b0-440b-b325-acb6da0bd50b Forscher hoffen auf Durchbruch für die Medikamentenforschung] (Researchers hope for a breakthrough for drug research), ''[[Der Spiegel]]'', 2 December 2020</ref> and ''[[The Spectator]]''.<ref>Bissan Al-Lazikani, [https://www.spectator.co.uk/article/the-solving-of-a-biological-mystery The solving of a biological mystery], ''[[The Spectator]]'', 1 December 2020</ref> In London ''[[The Times]]'' made the story its front-page photo lead, with two further pages of inside coverage and an editorial.<ref>Tom Whipple, "Deepmind computer solves new puzzle: life", ''[[The Times]]'', 1 December 2020. [https://twitter.com/UNSNUK/status/1333711800676835329 front page image], via Twitter.</ref><ref>Tom Whipple, [https://www.thetimes.co.uk/edition/news/deepmind-finds-biology-s-holy-grail-with-answer-to-protein-problem-htg6s7qlq Deepmind finds biology’s ‘holy grail’ with answer to protein problem], ''[[The Times]]'' (online), 30 November 2020.<br />In all science editor Tom Whipple wrote six articles on the subject for ''The Times'' on the day the news broke. ([https://twitter.com/whippletom/status/1333494448420958210 thread]).</ref> A frequent theme was that ability to predict protein structures accurately based on the constituent amino acid sequence is expected to have a wide variety of benefits in the life sciences space including accelerating advanced drug discovery and enabling better understanding of diseases.<ref name=":5">{{Cite journal|last=Callaway|first=Ewen|date=2020-11-30|title='It will change everything': DeepMind's AI makes gigantic leap in solving protein structures|url=https://www.nature.com/articles/d41586-020-03348-4|journal=Nature|language=en|doi=10.1038/d41586-020-03348-4}}</ref><ref>[[Tim Hubbard]], [https://timjph.medium.com/the-secret-of-life-part-2-the-solution-of-the-protein-folding-problem-c544f3a77ee3 The secret of life, part 2: the solution of the protein folding problem.], [[medium.com]], 30 November 2020</ref> |
Propelled by press releases from CASP and DeepMind,<ref name=CASP_release>[https://predictioncenter.org/casp14/doc/CASP14_press_release.html Artificial intelligence solution to a 50-year-old science challenge could ‘revolutionise’ medical research] (press release), [[CASP]] organising committee, 30 November 2020</ref><ref name=DeepMindAlpha2 /> AlphaFold 2's success received wide media attention.<ref>Brigitte Nerlich, [https://blogs.nottingham.ac.uk/makingsciencepublic/2020/12/04/protein-folding-and-science-communication-between-hype-and-humility/ Protein folding and science communication: Between hype and humility], [[University of Nottingham]] blog, 4 December 2020</ref> As well as news pieces in the specialist science press, such as ''[[Nature]]'',<ref name=":5" /> ''[[Science]]'',<ref name=":4" /> ''[[MIT Technology Review]]'',<ref name=":0" /> and ''[[New Scientist]]'',<ref>Michael Le Page, [https://www.newscientist.com/article/2261156-deepminds-ai-biologist-can-decipher-secrets-of-the-machinery-of-life/ DeepMind's AI biologist can decipher secrets of the machinery of life], ''[[New Scientist]]'', 30 November 2020</ref><ref>[https://www.newscientist.com/article/2261613-the-predictions-of-deepminds-latest-ai-could-revolutionise-medicine/ The predictions of DeepMind’s latest AI could revolutionise medicine], ''[[New Scientist]]'', 2 December 2020</ref> the story was widely covered by major national newspapers,<ref>[[Cade Metz]], [https://www.nytimes.com/2020/11/30/technology/deepmind-ai-protein-folding.html London A.I. Lab Claims Breakthrough That Could Accelerate Drug Discovery], ''[[New York Times]]'', 30 November 2020</ref><ref>Ian Sample,[https://www.theguardian.com/technology/2020/nov/30/deepmind-ai-cracks-50-year-old-problem-of-biology-research DeepMind AI cracks 50-year-old problem of protein folding], ''[[The Guardian]]'', 30 November 2020</ref><ref>Lizzie Roberts, [https://www.telegraph.co.uk/news/2020/11/30/google-ai-researchers-crack-50-year-old-protein-folding-problem/ 'Once in a generation advance' as Google AI researchers crack 50-year-old biological challenge]. ''[[Daily Telegraph]]'', 30 November 2020</ref><ref name=ElPais /> as well as general news-services and weekly publications, such as ''[[Fortune (magazine)|Fortune]]'',<ref>Jeremy Kahn, [https://fortune.com/2020/11/30/deepmind-protein-folding-breakthrough/ In a major scientific breakthrough, A.I. predicts the exact shape of proteins], ''[[Fortune (magazine)|Fortune]]'', 30 November 2020</ref><ref name=KahnLessons /> ''[[The Economist]]'',<ref name=":1" /> [[Bloomberg LP|Bloomberg]],<ref name=":2" /> ''[[Der Spiegel]]'',<ref name=Spiegel_1>Julia Merlot, [https://www.spiegel.de/wissenschaft/medizin/kuenstliche-intelligenz-sagt-faltung-von-proteinen-praezise-voraus-a-c52705ef-d3b0-440b-b325-acb6da0bd50b Forscher hoffen auf Durchbruch für die Medikamentenforschung] (Researchers hope for a breakthrough for drug research), ''[[Der Spiegel]]'', 2 December 2020</ref> and ''[[The Spectator]]''.<ref>Bissan Al-Lazikani, [https://www.spectator.co.uk/article/the-solving-of-a-biological-mystery The solving of a biological mystery], ''[[The Spectator]]'', 1 December 2020</ref> In London ''[[The Times]]'' made the story its front-page photo lead, with two further pages of inside coverage and an editorial.<ref>Tom Whipple, "Deepmind computer solves new puzzle: life", ''[[The Times]]'', 1 December 2020. [https://twitter.com/UNSNUK/status/1333711800676835329 front page image], via Twitter.</ref><ref>Tom Whipple, [https://www.thetimes.co.uk/edition/news/deepmind-finds-biology-s-holy-grail-with-answer-to-protein-problem-htg6s7qlq Deepmind finds biology’s ‘holy grail’ with answer to protein problem], ''[[The Times]]'' (online), 30 November 2020.<br />In all science editor Tom Whipple wrote six articles on the subject for ''The Times'' on the day the news broke. ([https://twitter.com/whippletom/status/1333494448420958210 thread]).</ref> A frequent theme was that ability to predict protein structures accurately based on the constituent amino acid sequence is expected to have a wide variety of benefits in the life sciences space including accelerating advanced drug discovery and enabling better understanding of diseases.<ref name=":5">{{Cite journal|last=Callaway|first=Ewen|date=2020-11-30|title='It will change everything': DeepMind's AI makes gigantic leap in solving protein structures|url=https://www.nature.com/articles/d41586-020-03348-4|journal=Nature|language=en|doi=10.1038/d41586-020-03348-4}}</ref><ref>[[Tim Hubbard]], [https://timjph.medium.com/the-secret-of-life-part-2-the-solution-of-the-protein-folding-problem-c544f3a77ee3 The secret of life, part 2: the solution of the protein folding problem.], [[medium.com]], 30 November 2020</ref> |
||
| Line 72: | Line 70: | ||
Finally, some have noted that even a perfect answer to the protein ''[[protein structure prediction|prediction]]'' problem would still leave questions about the protein ''[[protein folding|folding]]'' problem -- understanding in detail how the folding process actually occurs in nature (and how sometimes they can also [[proteopathy|misfold]]).<!-- eg https://twitter.com/daviddesancho/status/1333520127556546562 and other tweets, but there must be other references --> |
Finally, some have noted that even a perfect answer to the protein ''[[protein structure prediction|prediction]]'' problem would still leave questions about the protein ''[[protein folding|folding]]'' problem -- understanding in detail how the folding process actually occurs in nature (and how sometimes they can also [[proteopathy|misfold]]).<!-- eg https://twitter.com/daviddesancho/status/1333520127556546562 and other tweets, but there must be other references --> |
||
But even with such caveats about whether protein structure prediction could truly now be said to be a "solved" problem, there was universal recognition that the AlphaFold 2 results represented a huge technical step forward and intellectual achievement. |
But even with such caveats about whether protein structure prediction could truly now be said to be a "solved" problem, there was universal recognition that the AlphaFold 2 results represented a huge technical step forward and intellectual achievement.<ref>Cristina Sáez, [https://www.lavanguardia.com/ciencia/20201202/49848364758/alfonso-valencia-inteligencia-artificial-estructura-proteinas-deepmind.html El último avance fundamental de la biología se basa en la investigación de un científico español], ''[[La Vanguardia]]'', 2 December 2020</ref> |
||
== Applications == |
== Applications == |
||
Revision as of 01:30, 8 December 2020
AlphaFold is an artificial intelligence program developed by Google's DeepMind which performs predictions of protein structure.[1] The program is designed as a deep learning system that is built to predict folded protein structures to the width of an atom.[2]
In November 2020, a version of the program titled AlphaFold 2 took part in the 14th edition of the biennial Critical Assessment of Techniques for Protein Structure Prediction (CASP) competition,[3] in which it achieved a level of accuracy much higher than any other computational method.[2] The program scored above 90 for around two-thirds of the proteins in CASP's global distance test (GDT), a test that measures the degree to which a computational program predicted structure is similar to the lab experiment determined structure, with 100 being an exact match, within the distance cutoff used for calculating GDT.[2][4]
AlphaFold 2's results at CASP were described as "astounding",[5] and transformational.[6] Others have objected that any such statement is premature, given that AlphaFold's method has not yet been made known in sufficient detail to allow discussion, nor full assessment of its limitations, nor independent reimplementation; nor are the reasons for its success yet understood qualitiatively; and that considerable challenges remain before the questions of protein folding can be said to be fully understood. Nevertheless there has been widespread respect for the technical achievement.
Protein folding problem
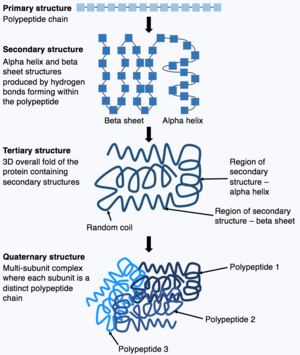
Proteins consist of chains of amino acids which spontaneously fold, in a process called protein folding, to form biologically important native state three dimensional structures. DNA sequences contain fundamental information about the sequences of these amino acids, but, the information about the protein folding and structures are determined by physical processes which can not be directly predicted from the DNA sequences.[7] Scientists look to experimental techniques such as X-ray crystallography, cryo-electron microscopy and nuclear magnetic resonance, which are both expensive and time-consuming to determine the target structures that proteins fold into.[7] Such efforts have identified the structures of about 170,000 proteins over the last 60 years, while there are over 200 million known proteins across life forms.[4] There are numerous computational methods of protein structure prediction, but their accuracy has not been close to experimental techniques, thus limiting their value.
Algorithm
While the details of AlphaFold's 2020 algorithms have not been publicly released, some are expected to be announced early December 2020, in the CASP conference. DeepMind is known to have trained the program on over 170,000 proteins from a public repository of protein sequences and structures. The program uses a form of attention network, a deep learning technique that focuses on having the AI algorithm solve parts of a larger problem and piece it together to obtain the overall solution.[2] The overall training was conducted on processing power between 100 and 200 GPUs.[2] Training the system on this hardware took "a few weeks", after which the program would take "a matter of days" to converge for each structure.[8]
AlphaFold 1 (2018) built on work developed by various teams in the 2010s, that looked at the large banks now available of related DNA sequences from many different organisms (mostly without known 3D structures), to try to find changes at different residues that appeared to be correlated, even though the residues were not consecutive in the main chain. Such correlations suggest that the residues may be close to each other physically, even though not close in the sequence, allowing a contact map to be estimated. Building on very recent work, AlphaFold 1 extended this to estimate a probability distribution for just how close the residues might be likely to be -- turning the contact map into a likely distance map; and also using more advanced learning methods than previously to develop the inference. Combining a potential based on this probability distribution with the calculated local free-energy of the configuration, the team were then able to use gradient descent to a solution that best fitted both.[9][10]
More technically, Torrisi et al. summarised the approach of AlphaFold version 1 as follows:[11]
Central to AlphaFold is a distance map predictor implemented as a very deep residual neural networks with 220 residual blocks processing a representation of dimensionality 64×64×128 – corresponding to input features calculated from two 64 amino acid fragments. Each residual block has three layers including a 3×3 dilated convolutional layer – the blocks cycle through dilation of values 1, 2, 4, and 8. In total the model has 21 million parameters. The network uses a combination of 1D and 2D inputs, including evolutionary profiles from different sources and co-evolution features. Alongside a distance map in the form of a very finely-grained histogram of distances, AlphaFold predicts Φ and Ψ angles for each residue which are used to create the initial predicted 3D structure. The AlphaFold authors concluded that the depth of the model, its large crop size, the large training set of roughly 29,000 proteins, modern Deep Learning techniques, and the richness of information from the predicted histogram of distances helped AlphaFold achieve a high contact map prediction precision.
Per the team at DeepMind, the current version of the program (AlphaFold 2) is significantly different from the original version that won CASP 13 in 2018.[12][13]
The team had identified that its previous approach, combining local physics with a guide potential derived from pattern recognition, had a tendency to over-account for interactions between residues that were nearby in the sequence compared to interactions between residues further apart along the chain. As a result, AlphaFold 1 had a tendency to prefer models with slightly more secondary structure (alpha helices and beta sheets) that was the case in reality (a form of overfitting).[14]
AlphaFold 1 contained a number of modules, each trained separately, that were used to produce the guide potential that was then combined with the physics-based energy potential. AlphaFold 2 replaced all of this with a system of sub-networks coupled together into a single differentiable end-to-end model, based entirely on pattern recognition, which was trained in an integrated way as a single integrated structure.[13][15] Local physics is applied only as a final refinement step, which only slightly adjusts the predicted structure.[14] A key part of the design are two modules, believed to be based on a transformer design, that effect a mathematical transformation of the relationship matrix between residue positions and other residue positions, and that between residue positions and different sequences in the sequence alignment of identified similar DNA sequences respectively.[15] These transformations have the effect of bringing relevant data together and filtering out irrelevant data for these two relationships, in a context-dependent way (the "attention mechanism"), that can itself be learnt from training data. Their output then informs the final prediction module.[15] As the trained system is iterated, these tend to first generate small clusters of amino acids, then ways to orient these clusters into an overall structure.[4]
The AlphaFold team believes that the newest version can be further developed, with room for further improvements in accuracy.[12]
Competitions

The crimson trend-line shows how a handful of models including AlphaFold 1 achieved a significant step-change in 2018 over the rate of progress that had previously been achieved, particularly in respect of the protein sequences considered the most difficult to predict.
(Qualitative improvement had been made in earlier years, but it is only as changes bring structures within 8 Å of their experimental positions that they start to affect the CASP GDS-TS measure).
The orange trend-line shows that by 2020 online prediction servers had been able to learn from and match this performance, while the best other groups (green curve) had on average been able to make some improvements on it. However, the black trend-curve shows the degree to which AlphaFold 2 had surpassed this again in 2020, across the board.
The detailed spread of data-points indicates the degree of consistency or variation achieved by AlphaFold. Outliers represent the handful of sequences for which it did not make such a successful prediction.
CASP13
In December 2018, DeepMind's AlphaFold was placed first in the overall rankings of the 13th Critical Assessment of Techniques for Protein Structure Prediction (CASP).
The program was particularly successfully predicting the most accurate structure for targets rated as the most difficult by the competition organisers, where no existing template structures were available from proteins with a partially similar sequence. AlphaFold gave the best prediction for 25 out of 43 protein targets in this class,[17][18][19] achieving a median score of 58.9 on the CASP's global distance test (GDT) score, ahead of 52.5 and 52.4 by the two next best-placed teams,[20] who were also using deep learning to estimate contact distances.[21][22] Overall, across all targets, the program achieved a GDT score of 68.5.[23]
In January 2020, the program code of AlphaFold 1 was released open-source on the source platform, GitHub.[24][7]
CASP14
In November 2020, DeepMind's new version, AlphaFold 2, won CASP14.[8][25] Overall, AlphaFold 2 made the best prediction for 88 out of the 97 targets.[5]
On the competition's preferred global distance test (GDT) measure of accuracy, the program achieved a median score of 92.4 (out of 100), meaning that more than half of its predictions were scored at better than 92.4% for having their atoms in more-or-less the right place,[26][27] a level of accuracy reported to be comparable to experimental techniques like X-ray crystallography.[12][6][23] In 2018 AlphaFold 1 had only reached this level of accuracy in two of all of its predictions.[5] 88% of predictions had a GDT-TS score of more than 80.[28]: slide 3 On the group of targets classed as the most difficult, AlphaFold 2 achieved a median score of 87.
Measured by the root-mean-square deviation (RMS-D) of the placement of the carbon atoms of the protein backbone chain, which tends to be dominated by the performance of the worst-fitted outliers, 88% of AlphaFold 2's predictions had an RMS deviation of less than 4 Å.[5] 76% of predictions achieved better than 3 Å, and 46% had an RMS accuracy better than 2 Å.[5] Overall the program achieved a median RMS deviation in its predictions of 2.1 Å.[5] For comparison, the bond length of a typical Carbon-Carbon bond is 1.5 Å. AlphaFold 2 also achieved an accuracy in modelling surface side chains described as "really really extraordinary".[16]: at 0:31:50
To additionally verify AlphaFold-2 the conference organisers approached four leading experimental groups for structures they were finding particularly challenging and had been unable to determine[28] In all four cases the three-dimensional models produced by AlphaFold 2 were sufficiently accurate to determine structures of these proteins by molecular replacement.[28] These included a cell membrane wedged protein, specifically a membrane protein from a species of Archaea microorganism, that the experimental team had been working on for ten years.[4]
Of the three structures that AlphaFold 2 had least success in predicting, two had been obtained by protein NMR methods,[16]: at 0:30:30 a setting that can lead to some different traits than the crystallographic structures that AlphaFold was mostly trained on. The third exists in nature as a protein complex consisting of 52 identical copies of the same protein (a "52-mer"),[16]: at 0:30:30 a situation AlphaFold was not programmed to consider. In such a structure a high contact probability between two residues could indicate a close proximity between particular residue positions from different copies of the protein, without requiring those positions to be near each other on a single copy of the protein, the only possibility allowed for in AlphaFold 2's single protein model. For all targets other than multimers, one target where only part of the sequence of a very large protein had been given for analysis, and the two structures determined by NMR, AlphaFold 2 achieved a GDT-TS score of over 80.[29]
Responses
AlphaFold 2 scoring more than 90 in CASP's global distance test (GDT) is considered a significant achievement in computational biology.[4] Nobel Prize winner and structural biologist Venki Ramakrishnan called the result "a stunning advance on the protein folding problem",[4] adding that "It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research."[8]
Propelled by press releases from CASP and DeepMind,[30][8] AlphaFold 2's success received wide media attention.[31] As well as news pieces in the specialist science press, such as Nature,[6] Science,[4] MIT Technology Review,[2] and New Scientist,[32][33] the story was widely covered by major national newspapers,[34][35][36][37] as well as general news-services and weekly publications, such as Fortune,[38][13] The Economist,[12] Bloomberg,[23] Der Spiegel,[39] and The Spectator.[40] In London The Times made the story its front-page photo lead, with two further pages of inside coverage and an editorial.[41][42] A frequent theme was that ability to predict protein structures accurately based on the constituent amino acid sequence is expected to have a wide variety of benefits in the life sciences space including accelerating advanced drug discovery and enabling better understanding of diseases.[6][43]
As summed up by Der Spiegel reservations about this coverage have focussed in two main areas: "There is still a lot to be done" and: "We don't even know how they do it".[44]
Although a 30-minute presentation about AlphaFold 2 was given on the second day of the CASP conference (December 1) by project leader John Jumper,[45] it appears to have been relatively limited on details. Neither slides nor a recording of the presentation are currently available from the CASP website.[46] DeepMind are expected to publish a scientific paper giving an account of AlphaFold 2 in the proceedings volume of the CASP conference; but it is not known whether it will go beyond what was said in the presentation.
Speaking to El País, Spanish researcher Alfonso Valencia said "The most important thing that this advance leaves us is knowing that this problem has a solution, that it is possible to solve it... We only know the result. Google does not provide the software and this is the frustrating part of the achievement because it will not directly benefit science."[37] Nevertheless as much as Google and DeepMind do release may help other teams develop similar AI systems, an "indirect" benefit.[37] In late 2019 DeepMind released much of the code of the first version of AlphaFold as open source; but only when work was well underway on the much more radical AlphaFold 2. It is not known whether it might do this again. Another option it could take might be to make AlphaFold 2 structure prediction available as an online black-box subscription service. Convergence for a single sequence has been estimated to require on the order of $10,000 worth of wholesale compute time.[47] But this would deny researchers access to the internal states of the system, the chance to learn more qualitatively what gives rise to AlphaFold 2's success, and the potential for new algorithms that could be lighter and more efficient yet still achieve such results. Fears of a potential for a lack of transparency by DeepMind have been contrasted with five decades of heavy public investment into the open Protein Data Bank and then also into open DNA sequence repositories, without which the data to train AlphaFold 2 would not have existed.[48][49][50]
In the immediate term AlphaFold 2 may prove most useful for a large number of structures, including membrane proteins, for which there is cryoEM or crystallographic data that it has not been possible to interpret. As was demonstrated at the conference, AlphaFold 2 may be able to produce predicted structures with sufficient accuracy and consistency to be biologically useful for a significantly wider range of targets than has so far been possible. AlphaFold 2's competition predictions showed unprecedented accuracy for proteins that stand by themselves. For such proteins AlphaFold 2's ability to predict structures for variant sequences may prove very useful in interpreting the effects of mutation and in protein design.
However it is not yet clear to what extent structure predictions made by AlphaFold 2 will hold up for proteins bound into complexes with other proteins and other molecules.[51] This was not a part of the CASP competition which AlphaFold entered, and not an eventuality it was internally designed to expect. Where structures that AlphaFold 2 did predict were for proteins that had strong interactions either with other copies of themselves, or with other structures, these were the cases where AlphaFold 2's predictions tended to be least refined and least reliable. As a large fraction of the most important biological machines in a cell comprise such complexes, or relate to how protein structures become modified when in contact with other molecules, this is an area that will continue to be the focus of considerable experimental attention.[51]
With so little yet known about the internal patterns that AF2 learns to make its predictions, it is not yet clear to what extent the program may be impaired in its ability to identify novel folds, if such folds are not well represented in the existing protein structures known in structure databases.[52][51] It is also not well known the extent to which protein structures in such databases, overwhelmingly of proteins that it has been possible to crystallise to X-ray, are representative of typical proteins that have not yet been crystallised. And it is also unclear how representative the frozen protein structures in crystals are of the dynamic structures found in the cells in vivo. AlphaFold 2's difficulties with structures obtained by protein NMR methods may not be a good sign.
In respect of drug discovery, Stephen Curry notes that while the resolution of AlphaFold 2's structures may be very good, the accuracy with which binding sites may need to be modelled may need to be even higher: typically 0.3 Å for molecular docking studies to be possible. So AlphaFold 2's structures may only be a limited help in such contexts.[52][51] Moreover, according to Science columnist Derek Lowe, because prediction of small-molecule binding even then is still not very good, computational prediction of drug targets is simply not in a position to take over as the "backbone" of corporate drug discovery -- so "protein structure determination simply isn’t a rate-limiting step in drug discovery in general".[53] Nevertheless, if better knowledge of protein structure could lead to better understanding of individual disease mechanisms and ultimately to better drug targets, or better understanding of the differences between human and animal models, ultimately that could lead to improvements.[54]
Finally, some have noted that even a perfect answer to the protein prediction problem would still leave questions about the protein folding problem -- understanding in detail how the folding process actually occurs in nature (and how sometimes they can also misfold).
But even with such caveats about whether protein structure prediction could truly now be said to be a "solved" problem, there was universal recognition that the AlphaFold 2 results represented a huge technical step forward and intellectual achievement.[55]
Applications
SARS-CoV-2
AlphaFold has been used to a predict structures of proteins of SARS-CoV-2, the causative agent of COVID-19. The structure of these proteins were pending experimental detection in early 2020.[56][6] Results were examined by the scientists at the Francis Crick Institute in the United Kingdom before release into the larger research community. The team also confirmed accurate prediction against the experimentally determined SARS-CoV-2 spike protein that was shared in the Protein Data Bank, an international open access database, before releasing the computationally determined structures of the under-studied protein molecules.[57] The team acknowledged that though these protein structures might not be the subject of ongoing therapeutical research efforts, they will add to the community's understanding of the SARS-CoV-2 virus.[57] Specifically, AlphaFold 2's prediction of the structure of the Orf3a protein was very similar to the structure determined by researchers at University of California, Berkeley using cryo-electron microscopy. This specific protein is believed to assist the virus in breaking out of the host cell once it replicates. This protein is also believed to play a role in triggering the inflammatory response to the infection.[58]
Published works
AlphaFold research
- Andrew W. Senior et al. (December 2019), "Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13)", Proteins: Structure, Function, Bioinformatics 87(12) 1141-1148 doi:10.1002/prot.25834
- Andrew W. Senior et al. (15 January 2020), "Improved protein structure prediction using potentials from deep learning", Nature 577 706–710 doi:10.1038/s41586-019-1923-7
- John Jumper et al. (December 2020), "High Accuracy Protein Structure Prediction Using Deep Learning", in Fourteenth Critical Assessment of Techniques for Protein Structure Prediction (Abstract Book), pp. 22–24
Derivative research
- Yang, Jianyi; Anishchenko, Ivan; Park, Hahnbeom; Peng, Zhenling; Ovchinnikov, Sergey; Baker, David (2019-11-18). "Improved protein structure prediction using predicted inter-residue orientations". bioRxiv: 846279. doi:10.1101/846279. S2CID 209563981.
- Billings, Wendy M.; Hedelius, Bryce; Millecam, Todd; Wingate, David; Corte, Dennis Della (2019-11-04). "ProSPr: Democratized Implementation of Alphafold Protein Distance Prediction Network". bioRxiv: 830273. doi:10.1101/830273. S2CID 209578310.
References
- ^ "AlphaFold". Deepmind. Retrieved 30 November 2020.
- ^ a b c d e f "DeepMind's protein-folding AI has solved a 50-year-old grand challenge of biology". MIT Technology Review. Retrieved 2020-11-30.
- ^ Shead, Sam (2020-11-30). "DeepMind solves 50-year-old 'grand challenge' with protein folding A.I." CNBC. Retrieved 2020-11-30.
- ^ a b c d e f g Robert F. Service, ‘The game has changed.’ AI triumphs at solving protein structures, Science, 30 November 2020
- ^ a b c d e f Mohammed AlQuraishi, CASP14 scores just came out and they’re astounding, twitter, 30 November 2020.
- ^ a b c d e Callaway, Ewen (2020-11-30). "'It will change everything': DeepMind's AI makes gigantic leap in solving protein structures". Nature. doi:10.1038/d41586-020-03348-4.
- ^ a b c "AlphaFold: Using AI for scientific discovery". Deepmind. Retrieved 2020-11-30.
- ^ a b c d e "AlphaFold: a solution to a 50-year-old grand challenge in biology". Deepmind. Retrieved 30 November 2020.
- ^ Mohammed AlQuraishi (May 2019), AlphaFold at CASP13, Bioinformatics, 35(22), 4862–4865 doi:10.1093/bioinformatics/btz422. See also Mohammed AlQuraishi (December 9, 2018), AlphaFold @ CASP13: “What just happened?” (blog post).
Mohammed AlQuraishi (15 January 2020), A watershed moment for protein structure prediction, Nature 577, 627-628 doi:10.1038/d41586-019-03951-0 - ^ AlphaFold: Machine learning for protein structure prediction, Foldit, 31 January 2020
- ^ Torrisi, Mirko et al. (22 Jan. 2020), Deep learning methods in protein structure prediction. Computational and Structural Biotechnology Journal vol. 18 1301-1310. doi:10.1016/j.csbj.2019.12.011 (CC-BY-4.0)
- ^ a b c d "DeepMind is answering one of biology's biggest challenges". The Economist. 2020-11-30. ISSN 0013-0613. Retrieved 2020-11-30.
- ^ a b c Jeremy Kahn, Lessons from DeepMind's breakthrough in protein-folding A.I., Fortune, 1 December 2020
- ^ a b John Jumper et al. (December 2020)
- ^ a b c See block diagram
- ^ a b c d John Moult (30 November 2020), CASP 14 introductory presentation, slide 19. See also CASP 14 video stream day 1 part 1, from 00:22:46
- ^ Sample, Ian (2 December 2018). "Google's DeepMind predicts 3D shapes of proteins". The Guardian. Retrieved 30 November 2020.
{{cite news}}: CS1 maint: url-status (link) - ^ "AlphaFold: Using AI for scientific discovery". Deepmind. Retrieved 30 November 2020.
- ^ Singh, Arunima (2020). "Deep learning 3D structures". Nature Methods. 17 (3): 249. doi:10.1038/s41592-020-0779-y. ISSN 1548-7105. PMID 32132733. S2CID 212403708.
- ^ See CASP 13 data tables for 043 A7D, 322 Zhang, and 089 MULTICOM
- ^ Wei Zheng et al,Deep-learning contact-map guided protein structure prediction in CASP13, Proteins: Structure, Function, and Bioinformatics, 87(12) 1149-1164 doi:10.1002/prot.25792; and slides
- ^ Jie Hou et al (2019), Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13, Proteins: Structure, Function, and Bioinformatics, 87(12) 1165-1178 doi:10.1002/prot.25697
- ^ a b c "DeepMind Breakthrough Helps to Solve How Diseases Invade Cells". Bloomberg.com. 2020-11-30. Retrieved 2020-11-30.
- ^ "deepmind/deepmind-research". GitHub. Retrieved 2020-11-30.
- ^ "DeepMind's protein-folding AI has solved a 50-year-old grand challenge of biology". MIT Technology Review. Retrieved 30 November 2020.
- ^ For the GDT-TS measure used, each atom in the prediction scores a quarter of a point if it is within 8 Å of the experimental position; half a point if it is within 4 Å, three-quarters of a point if it is within 2 Å, and a whole point if it is within 1 Å.
- ^ To achieve a GDT-TS score of 92.5, mathematically at least 70% of the structure must be accurate to within 1 Å, and at least 85% must be accurate to within 2 Å.
- ^ a b c Andriy Kryshtafovych (30 November 2020), Experimentalists: Are models useful? CASP 14 presentation. See also CASP 14 video stream day 1 part 1, from 0:34:30
- ^ Lisa Kinch et al, CASP14 Tertiary Structure Prediction Assessment:Topology (FM) Category (CASP 14 presentation), slide 11. See also CASP 14 video stream day 1 part 3, from 0:18:25
- ^ Artificial intelligence solution to a 50-year-old science challenge could ‘revolutionise’ medical research (press release), CASP organising committee, 30 November 2020
- ^ Brigitte Nerlich, Protein folding and science communication: Between hype and humility, University of Nottingham blog, 4 December 2020
- ^ Michael Le Page, DeepMind's AI biologist can decipher secrets of the machinery of life, New Scientist, 30 November 2020
- ^ The predictions of DeepMind’s latest AI could revolutionise medicine, New Scientist, 2 December 2020
- ^ Cade Metz, London A.I. Lab Claims Breakthrough That Could Accelerate Drug Discovery, New York Times, 30 November 2020
- ^ Ian Sample,DeepMind AI cracks 50-year-old problem of protein folding, The Guardian, 30 November 2020
- ^ Lizzie Roberts, 'Once in a generation advance' as Google AI researchers crack 50-year-old biological challenge. Daily Telegraph, 30 November 2020
- ^ a b c Nuño Dominguez, La inteligencia artificial arrasa en uno de los problemas más importantes de la biología (Artificial intelligence takes out one of the most important problems in biology), El País, 2 December 2020
- ^ Jeremy Kahn, In a major scientific breakthrough, A.I. predicts the exact shape of proteins, Fortune, 30 November 2020
- ^ Julia Merlot, Forscher hoffen auf Durchbruch für die Medikamentenforschung (Researchers hope for a breakthrough for drug research), Der Spiegel, 2 December 2020
- ^ Bissan Al-Lazikani, The solving of a biological mystery, The Spectator, 1 December 2020
- ^ Tom Whipple, "Deepmind computer solves new puzzle: life", The Times, 1 December 2020. front page image, via Twitter.
- ^ Tom Whipple, Deepmind finds biology’s ‘holy grail’ with answer to protein problem, The Times (online), 30 November 2020.
In all science editor Tom Whipple wrote six articles on the subject for The Times on the day the news broke. (thread). - ^ Tim Hubbard, The secret of life, part 2: the solution of the protein folding problem., medium.com, 30 November 2020
- ^ Christian Stöcker, Google greift nach dem Leben selbst (Google is reaching for life itself), Der Spiegel, 6 December 2020
- ^ CASP 14 meeting program. Accessed 7 December 2020.
- ^ CASP 14 website; as of 7 December 2020
- ^ Carlos Outeiral, CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics, Oxford Protein Informatics Group. (3 December)
- ^ Aled Edwards, The AlphaFold2 success: It took a village, via medium.com, 5 December 2020
- ^ David Briggs, If Google’s Alphafold2 really has solved the protein folding problem, they need to show their working, The Skeptic, 4 December 2020
- ^ The Guardian view on DeepMind’s brain: the shape of things to come, The Guardian, 6 December 2020
- ^ a b c d Tom Ireland, How will AlphaFold change bioscience research?, The Biologist, 4 December 2020
- ^ a b Stephen Curry, No, DeepMind has not solved protein folding, Reciprocal Space (blog), 2 December 2020
- ^ Derek Lowe, In the Pipeline: What’s Crucial And What Isn’t, Science Translational Medicine, 25 September 2019
- ^ Derek Lowe, In the Pipeline: The Big Problems, Science Translational Medicine, 1 December 2020
- ^ Cristina Sáez, El último avance fundamental de la biología se basa en la investigación de un científico español, La Vanguardia, 2 December 2020
- ^ "AI Can Help Scientists Find a Covid-19 Vaccine". Wired. ISSN 1059-1028. Retrieved 2020-12-01.
- ^ a b "Computational predictions of protein structures associated with COVID-19". Deepmind. Retrieved 2020-12-01.
- ^ "How DeepMind's new protein-folding A.I. is already helping to combat the coronavirus pandemic". Fortune. Retrieved 2020-12-01.
Further reading
- Carlos Outeiral, CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics, Oxford Protein Informatics Group. (3 December)
External links
- CASP 14 website
- AlphaFold: The making of a scientific breakthrough, DeepMind, via YouTube.
AlphaFold 1
- Senior, Andrew (August 23, 2019). "AlphaFold: improved protein structure prediction using potentials from deep learning". Institute for Protein Design – via YouTube.
- AlphaFold code used at CASP13 on GitHub
- Open source community implementation – ProSPr on GitHub
