Microbiota: Difference between revisions
Clarified the original definition of "microbiome," which was not used to describe the collective genomes of the microorganisms living in an environmental niche. Rather it was coined to describe the microorganisms themselves. |
minor error, changed an "in" to an "and" |
||
| Line 5: | Line 5: | ||
http://www.the-scientist.com/?articles.view/articleNo/13313/title/-Ome-Sweet--Omics---A-Genealogical-Treasury-of-Words/</ref><ref>The NIH HMP Working Group. 2009. The NIH Human Microbiome Project. Genome Res. 2009 December; 19(12): 2317–2323. |
http://www.the-scientist.com/?articles.view/articleNo/13313/title/-Ome-Sweet--Omics---A-Genealogical-Treasury-of-Words/</ref><ref>The NIH HMP Working Group. 2009. The NIH Human Microbiome Project. Genome Res. 2009 December; 19(12): 2317–2323. |
||
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2792171/</ref> This term was originally coined by [[Joshua Lederberg]], who argued the importance of microorganisms inhabiting the human body in health |
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2792171/</ref> This term was originally coined by [[Joshua Lederberg]], who argued the importance of microorganisms inhabiting the human body in health and disease. Many scientific articles distinguish "microbiome" and "microbiota" to describe either the collective genomes of the microorganisms that reside in an environmental niche or the microorganisms themselves, respectively.<ref>Backhed, F; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. 2005. Host-Bacterial Mutualism in the Human Intestine. Science 307, 1915 (2005);DOI: 10.1126/science.1104816 </ref><ref>Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. 2007. The Human Microbiome Project. Nature. 449:804-810. doi:10.1038/nature06244 </ref><ref>Ley, R.E.; Peterson, D.A.; Gordon, J.I. 2006. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. ''Cell''. 124: 837-848. DOI 10.1016/j.cell.2006.02.017 </ref> However by the original definitions these terms are synonymous. |
||
The human body contains over 10 times more microbial cells than human cells, although the entire microbiome only weighs about {{convert|200|g}},<ref>{{cite news|last=Zimmer|first=Carl|title=How Microbes Defend and Define Us|url=http://www.nytimes.com/2010/07/13/science/13micro.html?_r=2&pagewanted=all|accessdate=17 July 2010|newspaper=New York Times|date=13 July 2010}}</ref><ref>Because bacteria are [[Bacterial_cell_structure#The_importance_of_cell_size|10-100]] times smaller than human cells, the entire microbiome weighs about {{convert|200|g}}. See also {{Cite web |last=Coyle, MD |first=Walter J.| title=The Human Microbiome: The Undiscovered Country|page=16 |accessdate=2 March 2012 | url=http://www.acponline.org/about_acp/chapters/ca/microbiome.pptx |postscript=<!-- Bot inserted parameter. Either remove it; or change its value to "." for the cite to end in a ".", as necessary. -->{{inconsistent citations}}}}</ref> with some weight estimates ranging as high as 3 pounds (approximately 48 ounces or 1,400 grams). Some consider it to be a "newly discovered organ" since its existence was not generally recognized until the late 1990s and it is understood to potentially have overwhelming impact on human health. Modern DNA testing of their residues has enabled researchers to find the majority of these microbes, since the majority of them cannot be cultured in a lab using current techniques. Its most important aspect may be its possible effect on auto-immune diseases like diabetes, [[rheumatoid arthritis]], [[muscular dystrophy]], [[multiple sclerosis]], [[fibromyalgia]], and perhaps some [[cancer]]s. Common obesity might also be aggravated by a poor mix of microbes in the gut. Since some of the microbes in our body can modify the production of neurotransmitters known to be found in the brain, we may also find some relief for schizophrenia, depression, bipolar disorder and other neuro-chemical imbalances. |
The human body contains over 10 times more microbial cells than human cells, although the entire microbiome only weighs about {{convert|200|g}},<ref>{{cite news|last=Zimmer|first=Carl|title=How Microbes Defend and Define Us|url=http://www.nytimes.com/2010/07/13/science/13micro.html?_r=2&pagewanted=all|accessdate=17 July 2010|newspaper=New York Times|date=13 July 2010}}</ref><ref>Because bacteria are [[Bacterial_cell_structure#The_importance_of_cell_size|10-100]] times smaller than human cells, the entire microbiome weighs about {{convert|200|g}}. See also {{Cite web |last=Coyle, MD |first=Walter J.| title=The Human Microbiome: The Undiscovered Country|page=16 |accessdate=2 March 2012 | url=http://www.acponline.org/about_acp/chapters/ca/microbiome.pptx |postscript=<!-- Bot inserted parameter. Either remove it; or change its value to "." for the cite to end in a ".", as necessary. -->{{inconsistent citations}}}}</ref> with some weight estimates ranging as high as 3 pounds (approximately 48 ounces or 1,400 grams). Some consider it to be a "newly discovered organ" since its existence was not generally recognized until the late 1990s and it is understood to potentially have overwhelming impact on human health. Modern DNA testing of their residues has enabled researchers to find the majority of these microbes, since the majority of them cannot be cultured in a lab using current techniques. Its most important aspect may be its possible effect on auto-immune diseases like diabetes, [[rheumatoid arthritis]], [[muscular dystrophy]], [[multiple sclerosis]], [[fibromyalgia]], and perhaps some [[cancer]]s. Common obesity might also be aggravated by a poor mix of microbes in the gut. Since some of the microbes in our body can modify the production of neurotransmitters known to be found in the brain, we may also find some relief for schizophrenia, depression, bipolar disorder and other neuro-chemical imbalances. |
||
Revision as of 15:24, 7 August 2013
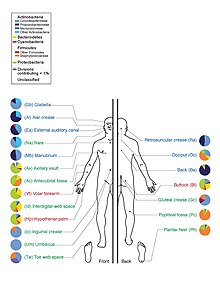
A microbiome is "the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space." [1][2] This term was originally coined by Joshua Lederberg, who argued the importance of microorganisms inhabiting the human body in health and disease. Many scientific articles distinguish "microbiome" and "microbiota" to describe either the collective genomes of the microorganisms that reside in an environmental niche or the microorganisms themselves, respectively.[3][4][5] However by the original definitions these terms are synonymous.
The human body contains over 10 times more microbial cells than human cells, although the entire microbiome only weighs about 200 grams (7.1 oz),[6][7] with some weight estimates ranging as high as 3 pounds (approximately 48 ounces or 1,400 grams). Some consider it to be a "newly discovered organ" since its existence was not generally recognized until the late 1990s and it is understood to potentially have overwhelming impact on human health. Modern DNA testing of their residues has enabled researchers to find the majority of these microbes, since the majority of them cannot be cultured in a lab using current techniques. Its most important aspect may be its possible effect on auto-immune diseases like diabetes, rheumatoid arthritis, muscular dystrophy, multiple sclerosis, fibromyalgia, and perhaps some cancers. Common obesity might also be aggravated by a poor mix of microbes in the gut. Since some of the microbes in our body can modify the production of neurotransmitters known to be found in the brain, we may also find some relief for schizophrenia, depression, bipolar disorder and other neuro-chemical imbalances.
Microbiomes are being characterized in many other environments as well, including soil, seawater and freshwater systems. It is believed that Endosymbiotic theory originally gave rise to more complex organisms, and continued to play a fundamental role in guiding their evolution and expansion into new niches.
To avoid confusion, it should be noted that the microbes being discussed are generally non-pathogenic (do not cause disease unless they grow abnormally), they exist in harmony and symbiotically with their hosts.[8]
Researchers have learned that much of the population of microbes found in the human body are not bacteria, but a very old class of single-celled organisms called archaea.[8]
Introduction
All plants and animals, from protists to humans, live in close association with microbial organisms, cf. the human microbiome. Up until relatively recently, however, the interactions of plants and animals with the microbial world have been defined mostly in the context of disease states and a relatively small number of symbiotic case studies. Organisms do not live in isolation, but have evolved in the context of complex communities. A number of advances have driven a change in this perception, which include, notably, the current ability to perform genomic and gene expression analyses of single cells and even entire microbial communities in the new disciplines of metagenomics and metatranscriptonomics, along with massive databases enabling this information to be accessible to researchers across multiple disciplines, and methods of mathematical analysis that enable sense to be made of complex data sets. It has become increasingly appreciated that microbes make up an important part of an organism's phenotype, far beyond the occasional symbiotic case study.[9]
An organism's complement of microbial inhabitants has been called a "forgotten organ".[10]
Case studies
The following sections present various case studies which illustrate this concept. There is a strengthening consensus among evolutionary biologists that one should not separate an organism's genes from the context of its resident microbes.
Studies in humans
- Community sequencing of total gut microbiota taken from obese and lean twins show substantial differences in their compositions. Total population sequences were analyzed to determine the levels of enzymes involved in carbohydrate, lipid, and amino acid metabolism. Obesity is associated with phylum-level differences in the microbiota, a significantly reduced bacterial diversity, and an increase in the population expression of enzymes which result in an increased efficiency of calorie harvest in the diets of the obese twins.[11]
- Type I diabetes is an autoimmune disease that is correlated with a multiplicity of predisposing factors, including an aberrant intestinal microbiota, a leaky intestinal mucosal barrier, and intrinsic differences in immune responsiveness. Various animal models for diabetes have shown a role for bacteria in the onset of the disease. Community DNA sequencing of intestinal flora comparing healthy and autoimmune children showed that autoimmune children had relatively unstable gut biomes with significantly decreased levels of species diversity, and the populations showed large scale replacement of Firmicutes species with Bacteroidetes species.[12]
- Human skin represents the most extensive organ of the human body, whose functions include protecting the body from pathogens, preventing loss of moisture, and participating in the regulation of body temperature. Considered as an ecosystem, the skin supports a range of microbial communities that live in distinct niches. Hair-covered scalp lies but a few inches from exposed neck, which in turn lies inches away from moist hairy underarms, but these niches are, at a microbial level, as distinct as a temperate forest would be compared with savanna and tropical rain forest. Studies characterizing the microbiota that inhabit these different niches are providing insights into the balance between skin health and disease.[13]
- Prevention of urogenital diseases in women depends on healthy vaginal microbiomes, but what is meant by "healthy" has not been understood. Community population studies using advanced sequencing methodologies (including pyrosequencing) are yielding insights into the range of microbial diversity in the human vagina. An unexpected finding was the prevalence of Prevotella species, which are known to positively affect the growth of Gardnerella vaginalis and Peptostreptococcus anaerobius, two species linked to bacterial vaginosis, by providing these disease-associated bacteria with key nutrients.[14]
- A proposal has been made to classify people by enterotype, based on the composition of the gut microbiome. By combining 22 newly sequenced fecal metagenomes of individuals from four countries with previously published data sets, three robust clusters were identified that are not nation or continent specific.[15][16]

- The traditional view of the immune system is that it is a complex assembly of organs, tissues, cells and molecules that work together to eliminate pathogens. Modifications to this traditional view, that the immune system has evolved to control microbes, have come from the discovery that microbes coevolve with and exert control upon the immune system. It is known that germ-free animals possess an underdeveloped immune system. The biology of the T helper 17 cells (Th17) has generated interest due to their key role in inflammatory processes. Excessive amounts of the cell are thought to play a key role in autoimmune diseases such as multiple sclerosis, psoriasis, juvenile diabetes, rheumatoid arthritis, Crohn's disease, and autoimmune uveitis. It has been discovered that specific microbiota direct the differentiation of Th17 cells in the mucosa of the small intestine.[17]
Animal studies

- A massive, worldwide decline in amphibian populations has been well-publicised. Habitat loss and over-exploitation account for part of the problem, but many other processes seem to be at work. The spread of the virulent fungal disease chytridiomycosis represents an enigma.[18] The ability of some species to coexist with the causative agent Batrachochytrium dendrobatidis appears to be due to the expression of antimicrobial skin peptides along with the presence of symbiotic microbes that benefit the host by resisting pathogen colonization or inhibiting their growth while being themselves resistant to high concentrations of antimicrobial skin peptides.[19]
- The bovine rumen harbors a complex microbiome that converts plant cell wall biomass into proteins, short chain fatty acids, and gases. Multiple species are involved in this conversion. Traditional methods of characterizing the microbial population, based on culture analysis, missed many of the participants in this process. Comparative metagenomic studies yielded the surprising result that individual steer had markedly different community structures, predicted phenotype, and metabolic potentials,[20] even though they were fed identical diets, were housed together, and were apparently functionally identical in their utilization of plant cell wall resources.
- Leaf-cutter ants form huge underground colonies with millions of workers, each colony harvesting hundreds of kilograms of leaves each year. Unable to digest the cellulose in the leaves directly, they maintain fungus gardens that are the colony's primary food source. The fungus itself does not digest cellulose. Instead, a microbial community containing a diversity of bacteria is responsible for cellulose digestion. Analysis of the microbial population's genomic content by community metagenome sequencing methods revealed the presence of many genes with a role in cellulose digestion. This microbiome's predicted carbohydrate-degrading enzyme profile is similar to that of the bovine rumen, but the species composition is almost entirely different.[21]
Plant studies

- Plants exhibit a broad range of relationships with symbiotic microorganisms, ranging from parasitism, in which the association is disadvantageous to the host organism, to mutualism, in which the association is beneficial to both, to commensalism, in which the symbiont benefits while the host is not affected. Exchange of nutrients between symbiotic partners is an important part of the relationship: it may be bidirectional or unidirectional, and it may be context dependent. The strategies for nutrient exchange are highly diverse. Oomycetes and fungi have, through convergent evolution, developed similar morphology and occupy similar ecological niches. They develop hyphae, filamentous structures that penetrate the host cell. In those cases where the association is mutualistic, the plant often exchanges hexose sugars for inorganic phosphate from the fungal symbiont. It is speculated that such associations, which are very ancient, may have aided plants when they first colonized land.[22][23]
- A huge range of bacterial symbionts colonize plants. Many of these are pathogenic, but others known as plant-growth promoting bacteria (PGPB) provide the host with essential services such as nitrogen fixation, solubilization of minerals such as phosphorus, synthesis of plant hormones, direct enhancement of mineral uptake, and protection from pathogens.[24][25] PGPBs may protect plants from pathogens by competing with the pathogen for an ecological niche or a substrate, producing inhibitory allelochemicals, or inducing systemic resistance in host plants to the pathogen[26]
Effects on cognition
Depression
Microbes are also implicated in depression. The pathogenic bacteria Borrelia burgdorferii causes Lyme disease which causes depression in up to 2/3 of all cases.[27] Non-pathogenic bacteria are also implicated in depression in which bacterial populations are suppressed. One model of depression is periodic separation of infant mice from their mothers. These mice show reductions in Lactobacillus and Bifidobacterium species, functional gut abnormalities, increased corticosterone (stress hormone) levels, weight loss, and causes them to not swim as much in a forced swim test as control mice indicating behavioural despair. Treating the mice with Lactobacillus lowered corticosterone levels and gut abnormalities.[28] Another experiment has replicated the effect that germ free mice have an exaggerated stress response and also found reduced expression of brain-derived neurotrophic factor in the cortex and hippocampus.[29] Another experiment showed that treating the maternally separated mice with a probiotic culture of Bifodobacterium infantis minimizes weight loss, causes mice to swim longer and causes an increase in the amount of the serotonin precursor tryptophan produced.[30] Increasing serotonin levels through selective serotonin reuptake inhibitors is the primary treatment of depression in humans. Human patients with depression are less able to properly digest fructose,[31] which is also associated with a reduction in tryptophan production.[32] Eliminating fructose from their diet improved their depression.[33]
Anxiety
Gut microbes are also implicated in anxiety disorders. In humans anxiety disorders are common in patients with disturbed gut flora.[34] The bacteria Campylobacter jejuni has been shown to cause anxious behaviour in mice.[35] Germ free mice show less anxious behaviour and also less NR2B mRNA expression selectively in the central amygdala which might be responsible for the anxiolytic behaviour since NR2B antagonists have an anxiolytic effect on behaviour.[36] The behavioural change might also be caused by increased brain derived neurotrophic factor (BDNF) mRNA expression possibly inducing plasticity in the dentate granular layer of the hippocampus.[37] BDNF and the hippocampus are implicated in memory. Increased gut bacterial diversity has been shown to improve both working and reference memory as well as reducing anxiety-like behaviour.[38]
Autism
Autistic populations have a unique microbiome consisting of more clostridial species.[39] Half of all autistic children with gastrointestinal dysfunction were found to have the bacteria Sutterella which was completely absent in non-autistic children with gastrointestinal dysfunction.[40] There is evidence that for some children with late-onset autism antibiotics can alleviate symptoms temporarily.[41]
Human microbiome
The human microbiome consists of about 100 trillion microbial cells, outnumbering human cells 10 to 1.[42] Thus it can significantly affect human physiology. For example, in healthy individuals the microbiota provide a wide range of metabolic functions that humans lack.[43] In diseased individuals altered microbiota are associated with diseases such as neonatal necrotizing enterocolitis,[44] inflammatory bowel disease[45] and vaginosis.[14] Thus studying the human microbiome is an important task that has been undertaken by initiatives such as the Human Microbiome Project[46] and MetaHIT.[47]
Studying the human microbiome

The problem of elucidating the human microbiome is essentially identifying the members of a microbial community which includes bacteria, eukaryotes, and viruses. This is done primarily using DNA-based studies, though RNA, protein and metabolite based studies are also performed.[48] DNA-based microbiome studies typically can be categorized as either targeted amplicon studies or more recently shotgun metagenomic studies. The former focuses on specific known marker genes and is primarily informative taxonomically, while the latter is an entire metagenomic approach which can also be used to study the functional potential of the community. One of the challenges that is present in human microbiome studies but not in other metagenomic studies is to avoid including the host DNA in the study.[49]
Presence of a core microbiome
Aside from simply elucidating the composition of the human microbiome, one of the major questions involving the human microbiome is whether there is a "core", that is, whether there is a subset of the community that is shared between most humans.[50][51] If there is a core, then it would be possible to associate certain community compositions with disease states, which is one of the goals of the Human Microbiome Project. It is known that the human microbiome is highly variable both within a single subject and between different individuals. For example, the gut microbiota of humans is markedly dissimilar between individuals, a phenomenon which is also observed in mice.[52] Hamady and Knight show that one can rule out the possibility that any species is shared among all humans at more than 0.9% abundance in the gut or at more than 2% abundance on hands.[51] Although there is very little species level conservation between individuals, it has been shown that this may be a result of functional redundancy as different communities tend to converge on the same functional state.[11]
On 13 June 2012, a major milestone of the Human Microbiome Project (HMP) was announced by the NIH director Francis Collins.[53] The announcement was accompanied with a series of coordinated articles published in Nature[54][55] and several journals in the Public Library of Science (PLoS) on the same day. By mapping the normal microbial make-up of healthy humans using genome sequencing techniques, the researchers of the HMP have created a reference database and the boundaries of normal microbial variation in humans. From 242 healthy U.S. volunteers, more than 5,000 samples were collected from tissues from 15 (men) to 18 (women) body sites such as mouth, nose, skin, lower intestine (stool), and vagina. All the DNA, human and microbial, were analyzed with DNA sequencing machines. The microbial genome data were extracted by identifying the bacterial specific ribosomal RNA, 16S rRNA. The researchers calculated that more than 10,000 microbial species occupy the human ecosystem and they have identified 81 – 99% of the genera.
Hologenome theory of evolution

The hologenome theory proposes that the object of natural selection is not the individual organism, but the organism together with its associated microbial communities.
The hologenome theory originated in studies on coral reefs. Coral reefs are the largest structures created by living organisms, and contain abundant and highly complex microbial communities. Over the past several decades, major declines in coral populations have occurred. Climate change, water pollution and over-fishing are three stress factors that have been described as leading to disease susceptibility. Over twenty different coral diseases have been described, but of these, only a handful have had their causative agents isolated and characterized. Coral bleaching is the most serious of these diseases. In the Mediterranean Sea, the bleaching of Oculina patagonica was first described in 1994 and shortly determined to be due to infection by Vibrio shiloi. From 1994 to 2002, bacterial bleaching of O. patagonica occurred every summer in the eastern Mediterranean. Surprisingly, however, after 2003, O. patagonica in the eastern Mediterranean has been resistant to V. shiloi infection, although other diseases still cause bleaching. The surprise stems from the knowledge that corals are long lived, with lifespans on the order of decades,[56] and do not have adaptive immune systems. Their innate immune systems do not produce antibodies, and they should seemingly not be able to respond to new challenges except over evolutionary time scales. The puzzle of how corals managed to acquire resistance to a specific pathogen led Eugene Rosenberg and Ilana Zilber-Rosenburg to propose the Coral Probiotic Hypothesis. This hypothesis proposes that a dynamic relationship exists between corals and their symbiotic microbial communities. By altering its composition, this "holobiont" can adapt to changing environmental conditions far more rapidly than by genetic mutation and selection alone. Extrapolating this hypothesis of adaptation and evolution to other organisms, including higher plants and animals, led to the proposal of the Hologenome Theory of Evolution.[57]
The hologenome theory is still being debated.[58] A major criticism has been the claim that V. shiloi was misidentified as the causative agent of coral bleaching, and that its presence in bleached O. patagonica was simply that of opportunistic colonization.[59] If this is true, the basic observation leading to the theory would be invalid. Nevertheless, the theory has gained significant popularity as a way of explaining rapid changes in adaptation that cannot otherwise be explained by traditional mechanisms of natural selection. For those who accept the hologenome theory, the holobiont has become the principal unit of natural selection.
Research methods
Targeted amplicon sequencing
Targeted amplicon sequencing relies on having some expectations about the composition of the community that is being studied. In target amplicon sequencing a phylogenetically informative marker is targeted for sequencing. Such a marker should be present in ideally all the expected organisms. It should also evolve in such a way that it is conserved enough that primers can target genes from a wide range of organisms while evolving quickly enough to allow for finer resolution at the taxonomic level. A common marker for human microbiome studies is the gene for bacterial 16S rRNA (i.e. "16S rDNA", the sequence of DNA which encodes the ribosomal RNA molecule).[48] Since ribosomes are present in all living organisms, using 16S rDNA allows for DNA to be amplified from many more organisms than if another marker were used. The 16S rDNA gene contains both slowly evolving regions and fast evolving regions; the former can be used to design broad primers while the latter allow for finer taxonomic distinction. However, species-level resolution is not typically possible using the 16S rDNA. Primer selection is an important step, as anything that cannot be targeted by the primer will not be amplified and thus will not be detected. Different sets of primers have been shown to amplify different taxonomic groups due to sequence variation.
Targeted studies of eukaryotic and viral communities are limited[60] and subject to the challenge of excluding host DNA from amplification and the reduced eukaryotic and viral biomass in the human microbiome.[49]
After the amplicons are sequenced, molecular phylogenetic methods are used to infer the composition of the microbial community. This is done by clustering the amplicons into operational taxonomic units (OTUs) and inferring phylogenetic relationships between the sequences. Due to the complexity of the data, distance measures such as UniFrac distances are usually defined between microbiome samples, and downstream multivariate methods are carried out on the distance matrices. An important point is that the scale of data is extensive, and further approaches must be taken to identify patterns from the available information. Tools used to analyze the data include VAMPS,[61] QIIME[62] and mothur.[63]
Metagenomic sequencing
Metagenomics is also used extensively for studying microbial communities.[11][47][64] In metagenomic sequencing, DNA is recovered directly from environmental samples in an untargeted manner with the goal of obtaining an unbiased sample from all genes of all members of the community. Recent studies use shotgun Sanger sequencing or pyrosequencing to recover the sequences of the reads.[65] The reads can then be assembled into contigs. To determine the phylogenetic identity of a sequence, it is compared to available full genome sequences using methods such as BLAST. One drawback of this approach is that many members of microbial communities do not have a representative sequenced genome.[48]
Despite the fact that metagenomics is limited by the availability of reference sequences, one significant advantage of metagenomics over targeted amplicon sequencing is that metagenomics data can elucidate the functional potential of the community DNA.[66][67] Targeted gene surveys cannot do this as they only reveal the phylogenetic relationship between the same gene from different organisms. Functional analysis is done by comparing the recovered sequences to databases of metagenomic annotations such as KEGG. The metabolic pathways that these genes are involved in can then be predicted with tools such as MG-RAST,[68] CAMERA[69] and IMG/M.[70]
RNA and protein-based approaches
Metatranscriptomics studies have been performed to study the gene expression of microbial communities through methods such as the pyrosequencing of extracted RNA.[71] Structure based studies have also identified non-coding RNAs (ncRNAs) such as ribozymes from microbiota.[72] Metaproteomics is a new approach that studies the proteins expressed by microbiota, giving insight into its functional potential.[73]
Projects
The Human Microbiome Project (HMP) is a United States National Institutes of Health initiative with the goal of identifying and characterizing the microorganisms which are found in association with both healthy and diseased humans (their microbial flora). Launched in 2008, it is a five-year project, best characterized as a feasibility study, and has a total budget of $115 million. The ultimate goal of this and similar NIH-sponsored microbiome projects is to test how changes in the human microbiome are associated with human health or disease.
The Earth Microbiome Project (EMP) is an initiative to collect natural samples and analyze the microbial community around the globe. Microbes are highly abundant, diverse and have an important role in the ecological system. Yet as of 2010[update], it was estimated that the total global environmental DNA sequencing effort had produced less than 1 percent of the total DNA found in a liter of seawater or a gram of soil,[74] and the specific interactions between microbes are largely unknown. The EMP aims to process as many as 200,000 samples in different biomes, generating a complete database of microbes on earth to characterize environments and ecosystems by microbial composition and interaction. Using these data, new ecological and evolutionary theories can be proposed and tested.[75]
Conclusion
Many more case studies exist than the few presented in this article, which illustrate the diverse interactions that have been shown to exist between macro organisms and their microbial inhabitants. Elucidation of these interactions has required new technologies and an interdisciplinary approach. Genomics and ecology, once separate disciplines, are showing rapid convergence, and may together allow us to understand the molecular basis underlying the adaptations and interactions of the communities of life.[9]
See also
References
- ^ Lederberg J, McCray AT. ’Ome Sweet ’Omics—a genealogical treasury of words. Scientist. 2001;15:8. http://www.the-scientist.com/?articles.view/articleNo/13313/title/-Ome-Sweet--Omics---A-Genealogical-Treasury-of-Words/
- ^ The NIH HMP Working Group. 2009. The NIH Human Microbiome Project. Genome Res. 2009 December; 19(12): 2317–2323. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2792171/
- ^ Backhed, F; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. 2005. Host-Bacterial Mutualism in the Human Intestine. Science 307, 1915 (2005);DOI: 10.1126/science.1104816
- ^ Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. 2007. The Human Microbiome Project. Nature. 449:804-810. doi:10.1038/nature06244
- ^ Ley, R.E.; Peterson, D.A.; Gordon, J.I. 2006. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 124: 837-848. DOI 10.1016/j.cell.2006.02.017
- ^ Zimmer, Carl (13 July 2010). "How Microbes Defend and Define Us". New York Times. Retrieved 17 July 2010.
- ^ Because bacteria are 10-100 times smaller than human cells, the entire microbiome weighs about 200 grams (7.1 oz). See also Coyle, MD, Walter J. "The Human Microbiome: The Undiscovered Country". p. 16. Retrieved 2 March 2012Template:Inconsistent citations
{{cite web}}: CS1 maint: postscript (link) - ^ a b Madigan, Michael T. (2012). Brock biology of microorganisms (13th ed.). San Francisco: Benjamin Cummings. ISBN 9780321649638.
- ^ a b Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/j.zool.2011.04.001, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/j.zool.2011.04.001instead. - ^ Willyard C (24 November 2011). "Microbiome: Gut reaction". Nature. 479 (7374): S5–S7. doi:10.1038/479S5a.
- ^ a b c Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19043404, please use {{cite journal}} with
|pmid=19043404instead. - ^ Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW (2011). "Toward defining the autoimmune microbiome for type 1 diabetes". The ISME Journal. 5 (1): 82–91. doi:10.1038/ismej.2010.92. PMC 3105672. PMID 20613793.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA (2009). "Topographical and Temporal Diversity of the Human Skin Microbiome". Science. 324 (5931): 1190–2. Bibcode:2009Sci...324.1190G. doi:10.1126/science.1171700. PMC 2805064. PMID 19478181.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20534435, please use {{cite journal}} with
|pmid=20534435instead. - ^ Zimmer, Carl (20 April 2011). "Bacteria Divide People Into 3 Types, Scientists Say". The New York Times. Retrieved 21 April 2011.
a group of scientists now report just three distinct ecosystems in the guts of people they have studied.
- ^ Arumugam, Manimozhiyan (2010). "Enterotypes of the human gut microbiome". Nature. 473 (7346): 174–80. Bibcode:2011Natur.473..174.. doi:10.1038/nature09944. PMID 21508958.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR (2008). "Specific microbiota direct the differentiation of Th17 cells in the mucosa of the small intestine". Cell Host Microbe. 4 (4): 337–349. doi:10.1016/j.chom.2008.09.009. PMC 2597589. PMID 18854238.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW (2004). "Status and Trends of Amphibian Declines and Extinctions Worldwide" (PDF). Science. 306 (5702): 1783–6. Bibcode:2004Sci...306.1783S. doi:10.1126/science.1103538. PMID 15486254.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Woodhams DC, Rollins-Smith LA, Alford RA, Simon MA, Harris RN (2007). "Innate immune defenses of amphibian skin: antimicrobial peptides and more". Animal Conservation. 10 (4): 425–8. doi:10.1111/j.1469-1795.2007.00150.x.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA (2009). "Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases". Proc. Natl. Acad. Sci. USA. 106 (6): 1948–53. Bibcode:2009PNAS..106.1948B. doi:10.1073/pnas.0806191105. PMC 2633212. PMID 19181843.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomás AA, Foster CE, Pauly M, Weimer PJ, Barry KW, Goodwin LA, Bouffard P, Li L, Osterberger J, Harkins TT, Slater SC, Donohue TJ, Currie CR (2010). Sonnenburg, Justin (ed.). "An Insect Herbivore Microbiome with High Plant Biomass-Degrading Capacity". PLoS Genet. 6 (9): e1001129. doi:10.1371/journal.pgen.1001129. PMC 2944797. PMID 20885794.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Remy W, Taylor TN, Hass H, Kerp H; Taylor; Hass; Kerp (1994). "Four hundred-million-year-old vesicular arbuscular mycorrhizae". Proc. Natl. Acad. Sci. USA. 91 (25): 11841–3. Bibcode:1994PNAS...9111841R. doi:10.1073/pnas.91.25.11841. PMC 45331. PMID 11607500.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chibucos MC, Tyler BM (2009). "Common themes in nutrient acquisition by plant symbiotic microbes, described by the Gene Ontology". BMC Microbiology. 9(Suppl 1): S6. doi:10.1186/1471-2180-9-S1-S6.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Kloepper, J. W (1993). "Plant growth-promoting rhizobacteria as biological control agents". In Metting, F. B., Jr (ed.). Soil microbial ecology: applications in agricultural and environmental management. New York: Marcel Dekker Inc. pp. 255–274. ISBN 0-8247-8737-4.
{{cite book}}: CS1 maint: multiple names: editors list (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/S1369-5266(00)00183-7, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/S1369-5266(00)00183-7instead. - ^ Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005). "Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects". Appl Environ Microbiol. 71 (9): 4951–9. doi:10.1128/AEM.71.9.4951-4959.2005. PMC 1214602. PMID 16151072.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 7943444, please use {{cite journal}} with
|pmid=7943444instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1136/gut.2006.117176, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1136/gut.2006.117176instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 15133062, please use {{cite journal}} with
|pmid=15133062instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20696216, please use {{cite journal}} with
|pmid=20696216instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 10961700, please use {{cite journal}} with
|pmid=10961700instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 11336160, please use {{cite journal}} with
|pmid=11336160instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 11099057, please use {{cite journal}} with
|pmid=11099057instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 17206706, please use {{cite journal}} with
|pmid=17206706instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 17920243, please use {{cite journal}} with
|pmid=17920243instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 11517276, please use {{cite journal}} with
|pmid=11517276instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 21054680, please use {{cite journal}} with
|pmid=21054680instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19135464, please use {{cite journal}} with
|pmid=19135464instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1086/341914, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1086/341914instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1128/mBio.00261-11, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1128/mBio.00261-11instead.
- ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 10921511, please use {{cite journal}} with
|pmid=10921511instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1146/annurev.mi.31.100177.000543, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1146/annurev.mi.31.100177.000543instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16741115, please use {{cite journal}} with
|pmid=16741115instead. - ^ Mitchum, Rob (2012-03-05). "Bacterial frontier could yield future cures: UChicago scientists find untapped potential in the "microbiome" of bacteria inside the human body" (Web feature). The University of Chicago. Retrieved 2013-02-26.
- ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 12594638, please use {{cite journal}} with
|pmid=12594638instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1101/gr.096651.109, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1101/gr.096651.109instead. - ^ a b Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20203603, please use {{cite journal}} with
|pmid=20203603instead. - ^ a b c Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 22179717, please use {{cite journal}} with
|pmid=22179717instead. - ^ a b Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 18638418, please use {{cite journal}} with
|pmid=18638418instead. - ^ Tap, Julien; Mondot, Stanislas.; Levenez, Florence; Pelletier, Eric; Caron, Christophe; Furet, Jean-Pierre; Ugarte, Edgardo; Munoz-Tamayo, Rafael; Le Paslier, Denis (2009). "Towards the human intestinal microbiota phylogenetic core". Environmental Microbiology. 11 (102): 2574–2584. doi:10.1111/j.1462-2920.2009.01982.x. PMID 19601958.
- ^ a b Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19383763, please use {{cite journal}} with
|pmid=19383763instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 16033867, please use {{cite journal}} with
|pmid=16033867instead. - ^ "NIH Human Microbiome Project defines normal bacterial makeup of the body". NIH News. 13 June 2012.
- ^ The Human Microbiome Project Consortium, Barbara A.; Nelson, Karen E.; Pop, Mihai; Creasy, Heather H.; Giglio, Michelle G.; Huttenhower, Curtis; Gevers, Dirk; Petrosino, Joseph F.; Abubucker, Sahar (2012). "A framework for human microbiome research". Nature. 486 (7402): 215–221. Bibcode:2012Natur.486..215T. doi:10.1038/nature11209. PMC 3377744. PMID 22699610.
- ^ The Human Microbiome Project Consortium, Curtis; Gevers, Dirk; Knight, Rob; Abubucker, Sahar; Badger, Jonathan H.; Chinwalla, Asif T.; Creasy, Heather H.; Earl, Ashlee M.; Fitzgerald, Michael G. (2012). "Structure, function and diversity of the healthy human microbiome". Nature. 486 (7402): 207–214. Bibcode:2012Natur.486..207T. doi:10.1038/nature11234. PMC 3564958. PMID 22699609.
- ^ Baird AH, Bhagooli R, Ralph PJ, Takahashi S (2009). "Coral bleaching: the role of the host" (PDF). Trends in Ecology and Evolution. 24 (1): 16–20. doi:10.1016/j.tree.2008.09.005. PMID 19022522.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007). "The role of microorganisms in coral health, disease and evolution" (PDF). Nature Reviews Microbiology. 5 (5): 355–362. doi:10.1038/nrmicro1635. PMID 17384666.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Leggat W, Ainsworth T, Bythell J, Dove S, Gates R, Hoegh-Guldberg O, Iglesias-Prieto R, Yellowlees D (2007). "The hologenome theory disregards the coral holobiont". Nature Reviews Microbiology. 5 (10): Online Correspondence. doi:10.1038/nrmicro1635-c1.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ainsworth TD, Fine M, Roff G, Hoegh-Guldberg O (2008). "Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica". The ISME Journal. 2 (1): 67–73. doi:10.1038/ismej.2007.88. PMID 18059488.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20602987, please use {{cite journal}} with
|pmid=20602987instead. - ^ "VAMPS: The Visualization and Analysis of Microbial Population Structures". Bay Paul Center, MBL, Woods Hole. Retrieved 11 March 2012.
- ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 20383131, please use {{cite journal}} with
|pmid=20383131instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19801464, please use {{cite journal}} with
|pmid=19801464instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1126/science.1107851, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1126/science.1107851instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1371/journal.pcbi.1000667, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1371/journal.pcbi.1000667instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19900971, please use {{cite journal}} with
|pmid=19900971instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19880382, please use {{cite journal}} with
|pmid=19880382instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 18803844, please use {{cite journal}} with
|pmid=18803844instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 21045053, please use {{cite journal}} with
|pmid=21045053instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 17932063, please use {{cite journal}} with
|pmid=17932063instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 19444216, please use {{cite journal}} with
|pmid=19444216instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 21257745, please use {{cite journal}} with
|pmid=21257745instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 17431707, please use {{cite journal}} with
|pmid=17431707instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.4056/sigs.1433550, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.4056/sigs.1433550instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1186/2042-5783-1-5, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1186/2042-5783-1-5instead.
External links
- Systems Biology & the Microbiome
- US NIH human microbiome project page
- The International Human Microbiome Consortium
- The CIHR Canadian Microbiome Initiative
- Microbiome.org — a microbiome wiki portal site.
- The Earth Microbiome Project
- Microbiology's World Wide Web — by Joshua Lederberg
