Signal recognition particle RNA
| RN7SL1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | RN7SL1, 7L1a, 7SL, RN7SL, RNSRP1, Signal recognition particle RNA, RNA, 7SL, cytoplasmic 1, RNA component of signal recognition particle 7SL1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 612177; GeneCards: RN7SL1; OMA:RN7SL1 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||

The signal recognition particle RNA, (also known as 7SL, 6S, ffs, or 4.5S RNA) is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. SRP-receptor is a protein that is embedded in a membrane, and which contains a transmembrane pore. When the SRP-ribosome complex binds to SRP-receptor, SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the SRP-receptor transmembrane pore.
In this way SRP directs the movement of proteins within the cell to bind with a transmembrane pore which allows the protein to cross the membrane to where it is needed. The RNA and protein components of this complex are highly conserved but do vary between the different kingdoms of life.
The common SINE family Alu probably originated from a 7SL RNA gene after deletion of a central sequence.[3]
The eukaryotic SRP consists of a 300-nucleotide 7S RNA and six proteins: SRPs 72, 68, 54, 19, 14, and 9. Archaeal SRP consists of a 7S RNA and homologues of the eukaryotic SRP19 and SRP54 proteins. Eukaryotic and archaeal 7S RNAs have very similar secondary structures.[4]
In most bacteria, the SRP consists of an RNA molecule (4.5S) and the Ffh protein (a homologue of the eukaryotic SRP54 protein). Some Gram-positive bacteria (e.g. Bacillus subtilis) have a longer eukaryote-like SRP RNA that includes an Alu domain.[5]
In eukaryotes and archaea, eight helical elements fold into the Alu and S domains, separated by a long linker region.[6][7] The Alu domain is thought to mediate the peptide chain elongation retardation function of the SRP.[6] The universally conserved helix which interacts with the SRP54 M domain mediates signal sequence recognition.[7][8] The SRP19-helix 6 complex is thought to be involved in SRP assembly and stabilises helix 8 for SRP54. binding[6] Humans have three functional SRP RNA genes, conveniently named RN7SL1, RN7SL2, and RN7SL3. The human genome in particular is known to contain a large amount of SRP RNA related sequence, including Alu repeats.[5]
Discovery
[edit]SRP RNA was first detected in avian and murine oncogenic RNA (ocorna) virus particles.[9] Subsequently, SRP RNA was found to be a stable component of uninfected HeLa cells where it associated with membrane and polysome fractions.[10][11] In 1980, cell biologists purified from canine pancreas an 11S "signal recognition protein" (fortuitously also abbreviated "SRP") which promoted the translocation of secretory proteins across the membrane of the endoplasmic reticulum.[12] It was then discovered that SRP contained an RNA component.[13] Comparing the SRP RNA genes from different species revealed helix 8 of the SRP RNA to be highly conserved in all domains of life.[14] The regions near the 5′- and 3′-ends of the mammalian SRP RNA are similar to the dominant Alu family of middle repetitive sequences of the human genome.[15] It is now understood that Alu DNA originated from SRP RNA by excision of the central SRP RNA-specific (S) fragment, followed by reverse transcription and integration into multiple sites of the human chromosomes.[3] SRP RNAs have been identified also in some organelles, for example in the plastid SRPs of many photosynthetic organisms,[16] and in the nuclear ribosomal internal transcribed spacer region of several ectomycorrhizal fungi.[17]
Transcription and processing
[edit]Eukaryotic SRP RNAs are transcribed from DNA by RNA polymerase III (Pol III).[18] RNA polymerase III also transcribes the genes for 5S ribosomal RNA, tRNA, 7SK RNA, and U6 spliceosomal RNA. The promoters of the human SRP RNA genes include elements located downstream of the transcriptional start site. Plant SRP RNA promoters contain an upstream stimulatory element (USE) and a TATA box.[citation needed] Yeast SRP RNA genes have a TATA box and additional intragenic promoter sequences (referred to as A- and B-blocks) which play a role in regulating transcription of the SRP gene by Pol III.[19] In the bacteria, genes are organized in operons and transcribed by RNA polymerase.[citation needed] The 5′-end of the small (4.5S) SRP RNA of many bacteria is cleaved by RNase P.[20] The ends of the Bacillus subtilis SRP RNA are processed by RNase III. So far, no SRP RNA introns have been observed.[citation needed]
Function
[edit]
Co-translational translocation
[edit]The SRP RNA is an integral part of the small and the large domain of the SRP. The function of the small domain is to delay protein translation until the ribosome-bound SRP has an opportunity to associate with the membrane-resident SRP receptor (SR). Within the large domain, the SRP RNA of the signal peptide-charged SRP promotes the hydrolysis of two guanosine triphosphate (GTP) molecules. This reaction releases the SRP from the SRP receptor and the ribosome, allowing translation to continue and the protein to enter the translocon.[21] The protein transverses the membrane co-translationally (during translation) and enters into another cellular compartment or the extracellular space. In eukaryotes, the target is the membrane of the endoplasmic reticulum (ER). In Archaea, SRP delivers proteins to the plasma membrane.[22] In the bacteria, SRP primarily incorporates proteins into the inner membrane.[23]
Post-translational transport
[edit]SRP participates also in the sorting of proteins after their synthesis has been completed (post-translational protein sorting). In eukaryotes, tail-anchored proteins possessing a hydrophobic insertion sequence at their C-terminus are delivered to the endoplasmic reticulum (ER) by the SRP.[24] Similarly, the SRP assists post-translationally in the import of nuclear-encoded proteins to the thylakoid membrane of chloroplasts.[25]
Structure
[edit]
In 2005, a nomenclature for all SRP RNAs proposed a numbering system of 12 helices. Helix sections are named with a lower case letter suffix (e.g. 5a). Insertions, or helix "branches" are given dotted numbers (e.g. 9.1 and 12.1).
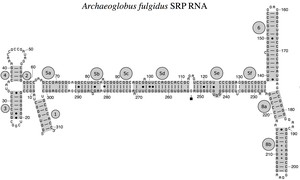
The SRP RNA spans a wide phylogenetic spectrum with respect to size and the number of its structural features (see the SRP RNA Secondary Structure Examples, below). The smallest functional SRP RNAs have been found in mycoplasma and related species. Escherichia coli SRP RNA (also called 4.5S RNA) is composed of 114 nucleotide residues and forms an RNA stem-loop. The gram-positive bacterium Bacillus subtilis encodes a larger 6S SRP RNA which resemble the Archaeal homologs but lacks SRP RNA helix 6. Archaeal SRP RNAs possess helices 1 to 8, lack helix 7, and are characterized by a tertiary structure which involves the apical loops of helix 3 and helix 4. The eukaryotic SRP RNAs lack helix 1 and contain a helix 7 of variable size. Some protozoan SRP RNAs have reduced helices 3 and 4. The ascomycota SRP RNAs have an altogether reduced small domain and lack helices 3 and 4. The largest SRP RNAs known to date are found in the yeasts (Saccharomycetes) which acquired helices 9 to 12 as insertions into helix 5, as well as an extended helix 7. Seed plants express numerous highly divergent SRP RNAs.[4]
Motifs
[edit]Four conserved features (motifs) have been identified (shown in the Figure in dark gray): the (1) SRP54 binding motif, (2) Helix 6 GNAR tetraloop motif, (3) 5e motif, and (4) UGU(NR) motif.[citation needed]
SRP54 binding
[edit]The asymmetric loop between helical sections 8a and 8b and the adjacent base paired 8b section are a prominent property of every SRP RNA. Helical section 8b contains non-Watson–Crick base pairings which contribute to the formation of a flatted minor groove in the RNA suitable for the binding of protein SRP54 (called Ffh in the bacteria).[7] The apical loop of helix 8 contains four, five, or six residues, depending on the species. It has a highly conserved guanosine as the first and an adenosine as the last loop residue. This feature is required for the interaction with the third adenosine residue of the helix 6 GNAR tetraloop motif.[26]
Helix 6 GNAR tetraloop
[edit]The SRP RNAs of eukaryotes and Archaea have a GNAR tetraloop (N is for any nucleotide, R is for a purine) in helix 6. Its conserved adenosine residue is important for the binding of protein SRP19.[27] This adenosine makes a tertiary interaction with another adenosine residue located in the apical loop of helix 8.[28]
5e
[edit]The 11 nucleotides of the 5e motif form four base pairs which are interrupted by a loop of three nucleotides.[5] In the eukaryotes, the first nucleotide of the loop is an adenosine which is needed for the binding of protein SRP72.[29]
UGU(NR)
[edit]The UGU(NR) motif connects helices 3 and 4 in the small (Alu) SRP domain. Fungal SRP RNAs lacking helices 3 and 4 contain the motif within the loop of helix 2.[5] It is important in the binding of the SRP9/14 protein heterodimer as part of an RNA U-turn.[30]
Secondary
[edit]-
Bacterial SRP RNA (6S RNA) from Bacillus subtilis
-
Archaeal SRP RNA Archaeoglobus fulgidus
-
Eukaryotic protist SRP RNA from Trypanosoma brucei
-
Eukaryotic yeast SRP RNA from Saccharomyces cerevisiae
Tertiary
[edit]| SRP RNA | |
|---|---|
| Identifiers | |
| Rfam | CL00003 |
| Other data | |
| PDB structures | PDBe 2IY3, 1Z43, 1RY1, 1QZW, 1MFQ, 1L9A, 1LNG,1JID, 1E8S, 1E8O, 1DUL, 1DUH,1D4R, 28SR, 28SP |
X-ray crystallography, nuclear magnetic resonance (NMR), and cryo-electron microscopy (cryo-EM] have been used to determine the molecular structure of portions of the SRP RNAs from various species. The available PDB structures show the RNA molecule either free or when bound to one or more SRP proteins.
-
SRP19-7S.S SRP RNA complex from M. jannaschii[28]
-
S-domain of human SRP[31]
Binding proteins
[edit]One or more SRP proteins bind to the SRP RNA to assemble the functional SRP. The SRP proteins are named according to their approximate molecular mass measured in kilodalton.[32] Most bacterial SRPs are composed of SRP RNA and SRP54 (also named Ffh for "Fifty-four homolog"). The Archaeal SRP contains proteins SRP54 and SRP19. In eukaryotes, the SRP RNA combines with the imported SRP proteins SRP9/14, SRP19, and SRP68/72 in a region of the nucleolus. This pre-SRP is transported to the cytosol where it binds to protein SRP54.[33] The molecular structures of the free or SRP RNA-bound proteins SRP9/14, SRP19, or SRP54 are known at high resolution.
SRP9 and SRP14
[edit]SRP9 and SRP14 are structurally related and form the SRP9/14 heterodimer which binds to the SRP RNA of the small (Alu) domain.[30] Yeast SRP lacks SRP9 and contains the structurally related binding protein SRP21. Yeast SRP14 forms homodimers in crystal and does not bind Alu.[34] SRP9/14 is absent in the SRP of trypanosoma which instead possess a tRNA-like molecule.[35]
SRP19
[edit]SRP19 is found in the SRP of eukaryotes and Archaea. Its primary role is in preparing the SRP RNA for the binding of SRP54, SRP68, and SRP72 by properly arranging SRP RNA helices 6 and 8.[31] Yeast SRP contains Sec65p, a larger homolog of SRP19.[36]
SRP54
[edit]Protein SRP54 (named Ffh in the bacteria) is an essential component of every SRP. It is composed of three functional domains: the N-terminal (N) domain, the GTPase (G) domain, and the methionine-rich (M) domain.[37][38]
SRP68 and SRP72
[edit]Proteins SRP68 and SRP72 are structurally unrelated constituents of the large domain of the eukaryotic SRP. They form a stable SRP68/72 heterodimer. About one third of the human SRP68 protein was shown to bind to the SRP RNA.[39] A relatively small region located near the C-terminus of SRP72 binds to the 5e SRP RNA motif.[29][40]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000276168 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Ullu E, Tschudi C (1984). "Alu sequences are processed 7SL RNA genes". Nature. 312 (5990): 171–172. Bibcode:1984Natur.312..171U. doi:10.1038/312171a0. PMID 6209580. S2CID 4328237.
- ^ a b Rosenblad MA, Larsen N, Samuelsson T, Zwieb C (2009). "Kinship in the SRP RNA family". RNA Biology. 6 (5): 508–516. doi:10.4161/rna.6.5.9753. PMID 19838050.
- ^ a b c d Regalia M, Rosenblad MA, Samuelsson T (August 2002). "Prediction of signal recognition particle RNA genes". Nucleic Acids Research. 30 (15): 3368–3377. doi:10.1093/nar/gkf468. PMC 137091. PMID 12140321.
- ^ a b c Wild K, Weichenrieder O, Strub K, Sinning I, Cusack S (February 2002). "Towards the structure of the mammalian signal recognition particle". Current Opinion in Structural Biology. 12 (1): 72–81. doi:10.1016/S0959-440X(02)00292-0. PMID 11839493.
- ^ a b c Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA (February 2000). "Crystal structure of the ribonucleoprotein core of the signal recognition particle". Science. 287 (5456): 1232–1239. Bibcode:2000Sci...287.1232B. doi:10.1126/science.287.5456.1232. PMID 10678824.
- ^ Batey RT, Sagar MB, Doudna JA (March 2001). "Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle". Journal of Molecular Biology. 307 (1): 229–246. doi:10.1006/jmbi.2000.4454. PMID 11243816.
- ^ Bishop JM, Levinson WE, Sullivan D, Fanshier L, Quintrell N, Jackson J (December 1970). "The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA". Virology. 42 (4): 927–937. doi:10.1016/0042-6822(70)90341-7. PMID 4321311.
- ^ Walker TA, Pace NR, Erikson RL, Erikson E, Behr F (September 1974). "The 7S RNA common to oncornaviruses and normal cells is associated with polyribosomes". Proceedings of the National Academy of Sciences of the United States of America. 71 (9): 3390–3394. Bibcode:1974PNAS...71.3390W. doi:10.1073/pnas.71.9.3390. PMC 433778. PMID 4530311.
- ^ Zieve G, Penman S (May 1976). "Small RNA species of the HeLa cell: metabolism and subcellular localization". Cell. 8 (1): 19–31. doi:10.1016/0092-8674(76)90181-1. PMID 954090. S2CID 26928799.
- ^ Walter P, Ibrahimi I, Blobel G (November 1981). "Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein". The Journal of Cell Biology. 91 (2 Pt 1): 545–550. doi:10.1083/jcb.91.2.545. PMC 2111968. PMID 7309795.
- ^ Walter P, Blobel G (October 1982). "Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum". Nature. 299 (5885): 691–698. Bibcode:1982Natur.299..691W. doi:10.1038/299691a0. PMID 6181418. S2CID 4237513.
- ^ Larsen N, Zwieb C (January 1991). "SRP-RNA sequence alignment and secondary structure". Nucleic Acids Research. 19 (2): 209–215. doi:10.1093/nar/19.2.209. PMC 333582. PMID 1707519.
- ^ Ullu E, Murphy S, Melli M (May 1982). "Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence". Cell. 29 (1): 195–202. doi:10.1016/0092-8674(82)90103-9. PMID 6179628. S2CID 12709599.
- ^ Rosenblad MA, Samuelsson T (November 2004). "Identification of chloroplast signal recognition particle RNA genes". Plant & Cell Physiology. 45 (11): 1633–1639. doi:10.1093/pcp/pch185. PMID 15574839.
- ^ Alm Rosenblad M, Martin MP, Tedersoo T, Ryberg MR, Larsson E, Wurzbacher C, Abarenkov K, Nilsson RH (2016). "Detection of signal recognition particle (SRP) RNAs in the nuclear ribosomal internal transcribed spacer 1 (ITS1) of three lineages of ectomycorrhizal fungi". MycoKeys (13): 21–33. doi:10.3897/mycokeys.13.8579. hdl:10261/163935.
- ^ Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (December 2007). "The expanding RNA polymerase III transcriptome". Trends in Genetics. 23 (12): 614–622. doi:10.1016/j.tig.2007.09.001. hdl:11381/1706964. PMID 17977614.
- ^ Dieci G, Giuliodori S, Catellani M, Percudani R, Ottonello S (March 2002). "Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae". The Journal of Biological Chemistry. 277 (9): 6903–6914. doi:10.1074/jbc.M105036200. PMID 11741971.
- ^ Bothwell, AL; Garber, RL; Altman, S (10 December 1976). "Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA". The Journal of Biological Chemistry. 251 (23): 7709–16. doi:10.1016/S0021-9258(17)32909-5. PMID 794064.
- ^ Shan SO, Walter P (February 2005). "Co-translational protein targeting by the signal recognition particle". FEBS Letters. 579 (4): 921–926. Bibcode:2005FEBSL.579..921S. doi:10.1016/j.febslet.2004.11.049. PMID 15680975. S2CID 46046514.
- ^ Zwieb C, Eichler J (March 2002). "Getting on target: the archaeal signal recognition particle". Archaea. 1 (1): 27–34. doi:10.1155/2002/729649. PMC 2685543. PMID 15803656.
- ^ Ulbrandt ND, Newitt JA, Bernstein HD (January 1997). "The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins". Cell. 88 (2): 187–196. doi:10.1016/S0092-8674(00)81839-5. PMID 9008159. S2CID 15246619.
- ^ Abell BM, Pool MR, Schlenker O, Sinning I, High S (July 2004). "Signal recognition particle mediates post-translational targeting in eukaryotes". The EMBO Journal. 23 (14): 2755–2764. doi:10.1038/sj.emboj.7600281. PMC 514945. PMID 15229647.
- ^ Schuenemann D, Gupta S, Persello-Cartieaux F, Klimyuk VI, Jones JD, Nussaume L, Hoffman NE (August 1998). "A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes". Proceedings of the National Academy of Sciences of the United States of America. 95 (17): 10312–10316. Bibcode:1998PNAS...9510312S. doi:10.1073/pnas.95.17.10312. PMC 21505. PMID 9707644.
- ^ Zwieb C, van Nues RW, Rosenblad MA, Brown JD, Samuelsson T (January 2005). "A nomenclature for all signal recognition particle RNAs". RNA. 11 (1): 7–13. doi:10.1261/rna.7203605. PMC 1370686. PMID 15611297.
- ^ Zwieb C (August 1992). "Recognition of a tetranucleotide loop of signal recognition particle RNA by protein SRP19". The Journal of Biological Chemistry. 267 (22): 15650–15656. doi:10.1016/S0021-9258(19)49585-9. PMID 1379233.
- ^ a b Hainzl T, Huang S, Sauer-Eriksson AE (June 2002). "Structure of the SRP19 RNA complex and implications for signal recognition particle assembly". Nature. 417 (6890): 767–771. Bibcode:2002Natur.417..767H. doi:10.1038/nature00768. PMID 12050674. S2CID 2509475.
- ^ a b Iakhiaeva E, Wower J, Wower IK, Zwieb C (June 2008). "The 5e motif of eukaryotic signal recognition particle RNA contains a conserved adenosine for the binding of SRP72". RNA. 14 (6): 1143–1153. doi:10.1261/rna.979508. PMC 2390789. PMID 18441046.
- ^ a b Weichenrieder O, Wild K, Strub K, Cusack S (November 2000). "Structure and assembly of the Alu domain of the mammalian signal recognition particle". Nature. 408 (6809): 167–173. Bibcode:2000Natur.408..167W. doi:10.1038/35041507. PMID 11089964. S2CID 4427070.
- ^ a b Kuglstatter A, Oubridge C, Nagai K (2002). "Induced structural changes of 7SL RNA during the assembly of human signal recognition particle". Nat Struct Biol. 9 (10): 740–744. doi:10.1038/nsb843. PMID 12244299. S2CID 9543041.
- ^ Walter P, Blobel G (September 1983). "Disassembly and reconstitution of signal recognition particle". Cell. 34 (2): 525–533. doi:10.1016/0092-8674(83)90385-9. PMID 6413076. S2CID 17907778.
- ^ Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T (January 2000). "Signal recognition particle components in the nucleolus". Proceedings of the National Academy of Sciences of the United States of America. 97 (1): 55–60. Bibcode:2000PNAS...97...55P. doi:10.1073/pnas.97.1.55. PMC 26615. PMID 10618370.
- ^ Brooks MA, Ravelli RB, McCarthy AA, Strub K, Cusack S (May 2009). "Structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle". Acta Crystallographica Section D. 65 (Pt 5): 421–433. doi:10.1107/S0907444909005484. PMID 19390147.
- ^ Liu L, Ben-Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S (May 2003). "The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule". The Journal of Biological Chemistry. 278 (20): 18271–18280. doi:10.1074/jbc.M209215200. PMID 12606550.
- ^ Hann BC, Stirling CJ, Walter P (April 1992). "SEC65 gene product is a subunit of the yeast signal recognition particle required for its integrity". Nature. 356 (6369): 532–533. Bibcode:1992Natur.356..532H. doi:10.1038/356532a0. PMID 1313947. S2CID 4287636.
- ^ Römisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B (August 1989). "Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains" (PDF). Nature. 340 (6233): 478–482. Bibcode:1989Natur.340..478R. doi:10.1038/340478a0. PMID 2502717. S2CID 4343347.
- ^ Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P (August 1989). "Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle". Nature. 340 (6233): 482–486. Bibcode:1989Natur.340..482B. doi:10.1038/340482a0. PMID 2502718. S2CID 619959.
- ^ Iakhiaeva E, Bhuiyan SH, Yin J, Zwieb C (June 2006). "Protein SRP68 of human signal recognition particle: identification of the RNA and SRP72 binding domains". Protein Science. 15 (6): 1290–1302. doi:10.1110/ps.051861406. PMC 2242529. PMID 16672232.
- ^ Iakhiaeva E, Yin J, Zwieb C (January 2005). "Identification of an RNA-binding domain in human SRP72". Journal of Molecular Biology. 345 (4): 659–666. doi:10.1016/j.jmb.2004.10.087. PMID 15588816.
Further reading
[edit]- Kuglstatter A, Oubridge C, Nagai K (October 2002). "Induced structural changes of 7SL RNA during the assembly of human signal recognition particle". Nature Structural Biology. 9 (10): 740–744. doi:10.1038/nsb843. PMID 12244299. S2CID 9543041.
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF (December 2007). "7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G". Journal of Virology. 81 (23): 13112–13124. doi:10.1128/JVI.00892-07. PMC 2169093. PMID 17881443.
- Ullu E, Weiner AM (December 1984). "Human genes and pseudogenes for the 7SL RNA component of signal recognition particle". The EMBO Journal. 3 (13): 3303–3310. doi:10.1002/j.1460-2075.1984.tb02294.x. PMC 557853. PMID 6084597.
- Englert M, Felis M, Junker V, Beier H (December 2004). "Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes". Biochimie. 86 (12): 867–874. doi:10.1016/j.biochi.2004.10.012. PMID 15667936.
- Oubridge C, Kuglstatter A, Jovine L, Nagai K (June 2002). "Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle". Molecular Cell. 9 (6): 1251–1261. doi:10.1016/S1097-2765(02)00530-0. PMID 12086622.
- Wang T, Tian C, Zhang W, Sarkis PT, Yu XF (January 2008). "Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging". Journal of Molecular Biology. 375 (4): 1098–1112. doi:10.1016/j.jmb.2007.11.017. PMID 18067920.
External links
[edit]- The SRP Database (SRPDB): Alignments of SRP RNAs and associated proteins, SRP RNA secondary structures and 3-D models
- Rfam entry for Metazoan type signal recognition particle RNA
- Rfam entry for Bacterial small signal recognition particle RNA
- Rfam entry for Bacterial large signal recognition particle RNA
- Rfam entry for Fungal signal recognition particle RNA
- Rfam entry for Plant signal recognition particle RNA
- Rfam entry for Protozoan signal recognition particle RNA
- Rfam entry for Archaeal signal recognition particle RNA
- Dnatube Signal Recognition Particle Movie







![SRP19-7S.S SRP RNA complex from M. jannaschii[28]](http://upload.wikimedia.org/wikipedia/commons/thumb/1/16/PDB_1lng_EBI.jpg/267px-PDB_1lng_EBI.jpg)
![S-domain of human SRP[31]](http://upload.wikimedia.org/wikipedia/commons/thumb/0/08/PDB_1mfq_EBI.jpg/267px-PDB_1mfq_EBI.jpg)