Binding site

In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity.[1] The binding partner of the macromolecule is often referred to as a ligand.[2] Ligands may include other proteins (resulting in a protein-protein interaction),[3] enzyme substrates,[4] second messengers, hormones, or allosteric modulators.[5] The binding event is often, but not always, accompanied by a conformational change that alters the protein's function.[6] Binding to protein binding sites is most often reversible (transient and non-covalent), but can also be covalent reversible[7] or irreversible.[8]
Function
Binding of a ligand to a binding site on protein often triggers a change in conformation in the protein and results in altered cellular function. Hence binding site on protein are critical parts of signal transduction pathways.[9] Types of ligands include neurotransmitters, toxins, neuropeptides, and steroid hormones.[10] Binding sites incur functional changes in a number of contexts, including enzyme catalysis, molecular pathway signaling, homeostatic regulation, and physiological function. Electric charge, steric shape and geometry of the site selectively allow for highly specific ligands to bind, activating a particular cascade of cellular interactions the protein is responsible for.[11][12]
Catalysis

Enzymes incur catalysis by binding more strongly to transition states than substrates and products. At the catalytic binding site, several different interactions may act upon the substrate. These range from electric catalysis, acid and base catalysis, covalent catalysis, and metal ion catalysis.[10] These interactions decrease the activation energy of a chemical reaction by providing favorable interactions to stabilize the high energy molecule. Enzyme binding allows for closer proximity and exclusion of substances irrelevant to the reaction. Side reactions are also discouraged by this specific binding.[13][10]
Types of enzymes that can perform these actions include oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases.[14]
For instance, the transferase hexokinase catalyzes the phosphorylation of glucose to make glucose-6-phosphate. Active site residues of hexokinase allow for stabilization of the glucose molecule in the active site and spur the onset of an alternative pathway of favorable interactions, decreasing the activation energy.[15]
Inhibition
Protein inhibition by inhibitor binding may induce obstruction in pathway regulation, homeostatic regulation and physiological function.
Competitive inhibitors compete with substrate to bind to free enzymes at active sites and thus impede the production of the enzyme-substrate complex upon binding. For example, carbon monoxide poisoning is caused by the competitive binding of carbon monoxide as opposed to oxygen in hemoglobin.
Uncompetitive inhibitors, alternatively, bind concurrently with substrate at active sites. Upon binding to an enzyme substrate (ES) complex, an enzyme substrate inhibitor (ESI) complex is formed. Similar to competitive inhibitors, the rate at product formation is decreased also.[4]
Lastly, mixed inhibitors are able to bind to both the free enzyme and the enzyme-substrate complex. However, in contrast to competitive and uncompetitive inhibitors, mixed inhibitors bind to the allosteric site. Allosteric binding induces conformational changes that may increase the protein's affinity for substrate. This phenomenon is called positive modulation. Conversely, allosteric binding that decreases the protein's affinity for substrate is negative modulation.[16]
Types
Active site
At the active site, a substrate binds to an enzyme to induce a chemical reaction.[17][18] Substrates, transition states, and products can bind to the active site, as well as any competitive inhibitors.[17] For example, in the context of protein function, the binding of calcium to troponin in muscle cells can induce a conformational change in troponin. This allows for tropomyosin to expose the actin-myosin binding site to which the myosin head binds to form a cross-bridge and induce a muscle contraction.[19]
In the context of the blood, an example of competitive binding is carbon monoxide which competes with oxygen for the active site on heme. Carbon monoxide's high affinity may outcompete oxygen in the presence of low oxygen concentration. In these circumstances, the binding of carbon monoxide induces a conformation change that discourages heme from binding to oxygen, resulting in carbon monoxide poisoning.[4]

Allosteric site
At the regulatory site, the binding of a ligand may elicit amplified or inhibited protein function.[4][20] The binding of a ligand to an allosteric site of a multimeric enzyme often induces positive cooperativity, that is the binding of one substrate induces a favorable conformation change and increases the enzyme's likelihood to bind to a second substrate.[21] Regulatory site ligands can involve homotropic and heterotropic ligands, in which single or multiple types of molecule affects enzyme activity respectively.[22]
Enzymes that are highly regulated are often essential in metabolic pathways. For example, phosphofructokinase (PFK), which phosphorylates fructose in glycolysis, is largely regulated by ATP. Its regulation in glycolysis is imperative because it is the committing and rate limiting step of the pathway. PFK also controls the amount of glucose designated to form ATP through the catabolic pathway. Therefore, at sufficient levels of ATP, PFK is allosterically inhibited by ATP. This regulation efficiently conserves glucose reserves, which may be needed for other pathways. Citrate, an intermediate of the citric acid cycle, also works as an allosteric regulator of PFK.[22][23]
Single- and multi-chain binding sites
Binding sites can be characterized also by their structural features. Single-chain sites (of “monodesmic” ligands, μόνος: single, δεσμός: binding) are formed by a single protein chain, while multi-chain sites (of "polydesmic” ligands, πολοί: many) [24] are frequent in protein complexes, and are formed by ligands that bind more than one protein chain, typically in or near protein interfaces. Recent research shows that binding site structure has profound consequences for the biology of protein complexes (evolution of function, allostery).[25][26]
Cryptic binding sites
Cryptic binding sites are the binding sites that are transiently formed in an apo form or that are induced by ligand binding. Considering the cryptic binding sites increases the size of the potentially “druggable” human proteome from ~40% to ~78% of disease-associated proteins.[27] The binding sites have been investigated by: support vector machine applied to "CryptoSite" data set,[27] Extension of "CryptoSite" data set,[28] long timescale molecular dynamics simulation with Markov state model and with biophysical experiments,[29] and cryptic-site index that is based on relative accessible surface area.[30]
Binding curves

Binding curves describe the binding behavior of ligand to a protein. Curves can be characterized by their shape, sigmoidal or hyperbolic, which reflect whether or not the protein exhibits cooperative or noncooperative binding behavior respectively.[31] Typically, the x-axis describes the concentration of ligand and the y-axis describes the fractional saturation of ligands bound to all available binding sites.[4] The Michaelis Menten equation is usually used when determining the shape of the curve. The Michaelis Menten equation is derived based on steady-state conditions and accounts for the enzyme reactions taking place in a solution. However, when the reaction takes place while the enzyme is bound to a substrate, the kinetics play out differently.[32]
Modeling with binding curves are useful when evaluating the binding affinities of oxygen to hemoglobin and myoglobin in the blood. Hemoglobin, which has four heme groups, exhibits cooperative binding. This means that the binding of oxygen to a heme group on hemoglobin induces a favorable conformation change that allows for increased binding favorability of oxygen for the next heme groups. In these circumstances, the binding curve of hemoglobin will be sigmoidal due to its increased binding favorability for oxygen. Since myoglobin has only one heme group, it exhibits noncooperative binding which is hyperbolic on a binding curve.[33]
Applications
Biochemical differences between different organisms and humans are useful for drug development. For instance, penicillin kills bacterial enzymes by inhibiting DD-transpeptidase, destroying the development of the bacterial cell wall and inducing cell death. Thus, the study of binding sites is relevant to many fields of research, including cancer mechanisms,[34] drug formulation,[35] and physiological regulation.[36] The formulation of an inhibitor to mute a protein's function is a common form of pharmaceutical therapy.[37]
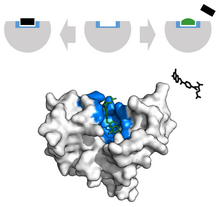
In the scope of cancer, ligands that are edited to have a similar appearance to the natural ligand are used to inhibit tumor growth. For example, Methotrexate, a chemotherapeutic, acts as a competitive inhibitor at the dihydrofolate reductase active site.[38] This interaction inhibits the synthesis of tetrahydrofolate, shutting off production of DNA, RNA and proteins.[38] Inhibition of this function represses neoplastic growth and improves severe psoriasis and adult rheumatoid arthritis.[37]
In cardiovascular illnesses, drugs such as beta blockers are used to treat patients with hypertension. Beta blockers (β-Blockers) are antihypertensive agents that block the binding of the hormones adrenaline and noradrenaline to β1 and β2 receptors in the heart and blood vessels. These receptors normally mediate the sympathetic "fight or flight" response, causing constriction of the blood vessels.[39]
Competitive inhibitors are also largely found commercially. Botulinum toxin, known commercially as Botox, is a neurotoxin causes flaccid paralysis in the muscle due to binding to acetylcholine dependent nerves. This interaction inhibits muscle contractions, giving the appearance of smooth muscle.[40]
Prediction
A number of computational tools have been developed for the prediction of the location of binding sites on proteins.[20][41][42] These can be broadly classified into sequence based or structure based.[42] Sequence based methods rely on the assumption that the sequences of functionally conserved portions of proteins such as binding site are conserved. Structure based methods require the 3D structure of the protein. These methods in turn can be subdivided into template and pocket based methods.[42] Template based methods search for 3D similarities between the target protein and proteins with known binding sites. The pocket based methods search for concave surfaces or buried pockets in the target protein that possess features such as hydrophobicity and hydrogen bonding capacity that would allow them to bind ligands with high affinity.[42] Even though the term pocket is used here, similar methods can be used to predict binding sites used in protein-protein interactions that are usually more planar, not in pockets.[43]
References
- ^ "Binding site". Medical Subject Headings (MeSH). U.S. National Library of Medicine.
The parts of a macromolecule that directly participate in its specific combination with another molecule.
- ^ "Ligands". Medical Subject Headings (MeSH). U.S. National Library of Medicine.
A molecule that binds to another molecule, used especially to refer to a small molecule that binds specifically to a larger molecule.
- ^ Amos-Binks A, Patulea C, Pitre S, Schoenrock A, Gui Y, Green JR, Golshani A, Dehne F (June 2011). "Binding site prediction for protein-protein interactions and novel motif discovery using re-occurring polypeptide sequences". BMC Bioinformatics. 12: 225. doi:10.1186/1471-2105-12-225. PMC 3120708. PMID 21635751.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Hardin CC, Knopp JA (2013). "Chapter 8: Enzymes". Biochemistry - Essential Concepts. New York: Oxford University Press. pp. 51–69. ISBN 978-1-62870-176-0.
- ^ Kenakin TP (April 2016). "Characteristics of Allosterism in Drug Action". In Bowery NG (ed.). Allosteric Receptor Modulation in Drug Targeting. CRC Press. p. 26. ISBN 978-1-4200-1618-5.
- ^ Spitzer R, Cleves AE, Varela R, Jain AN (April 2014). "Protein function annotation by local binding site surface similarity". Proteins. 82 (4): 679–94. doi:10.1002/prot.24450. PMC 3949165. PMID 24166661.
- ^ Bandyopadhyay A, Gao J (October 2016). "Targeting biomolecules with reversible covalent chemistry". Current Opinion in Chemical Biology. 34: 110–116. doi:10.1016/j.cbpa.2016.08.011. PMC 5107367. PMID 27599186.
- ^ Bellelli A, Carey J (January 2018). "Reversible Ligand Binding". Reversible Ligand Binding: Theory and Experiment. John Wiley & Sons. p. 278. ISBN 978-1-119-23848-5.
- ^ Xu D, Jalal SI, Sledge GW, Meroueh SO (October 2016). "Small-molecule binding sites to explore protein-protein interactions in the cancer proteome". Molecular BioSystems. 12 (10): 3067–87. doi:10.1039/c6mb00231e. PMC 5030169. PMID 27452673.
- ^ a b c Wilson K (March 2010). Principles and Techniques of Biochemistry and Molecular Biology. Cambridge University Press. pp. 581–624. doi:10.1017/cbo9780511841477.016. ISBN 9780511841477.
- ^ Ahern K (2015). Biochemistry Free For All. Oregon State University. pp. 110–141.
- ^ Kumar AP, Lukman S (2018-06-06). "Allosteric binding sites in Rab11 for potential drug candidates". PLOS One. 13 (6): e0198632. doi:10.1371/journal.pone.0198632. PMC 5991966. PMID 29874286.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Dobson JA, Gerrard AJ, Pratt JA (2008). Foundations of chemical biology. Oxford University Press. ISBN 9780199248995. OCLC 487962823.
- ^ Azzaroni O, Szleifer I (2017-12-04). Polymer and Biopolymer Brushes. doi:10.1002/9781119455042. ISBN 978-1-119-45501-1.
- ^ Dictionary of Food Science and Technology (2nd Edition). International Food Information Service. 2009. ISBN 978-1-4051-8740-4.
- ^ Clarke KG (2013). Bioprocess engineering. Woodhead Publishing. pp. 79–84. doi:10.1533/9781782421689. ISBN 978-1-78242-167-2.
- ^ a b Wilson K (March 2010). "Enzymes". In Wilson K, Walker J (eds.). Principles and Techniques of Biochemistry and Molecular Biology. Cambridge University Press. pp. 581–624. doi:10.1017/cbo9780511841477.016. ISBN 9780511841477. Retrieved 2018-11-01.
- ^ Schaschke C (2014). Dictionary of Chemical Engineering. Oxford University Press. ISBN 978-1-62870-844-8.
- ^ Morris J (2016). Biology How Life Works. United States of America: W.H. Freeman and Company. pp. 787–792. ISBN 978-1-4641-2609-3.
- ^ a b Konc J, Janežič D (April 2014). "Binding site comparison for function prediction and pharmaceutical discovery". Current Opinion in Structural Biology. 25: 34–9. doi:10.1016/j.sbi.2013.11.012. PMID 24878342.
- ^ Fuqua C, White D (2004). Prokaryotic Intercellular Signalling. Springer Netherlands. pp. 27–71. doi:10.1007/978-94-017-0998-9_2. ISBN 9789048164837.
{{cite book}}:|work=ignored (help) - ^ a b Creighton TE (2010). The Biophysical Chemistry of Nucleic Acids & Proteins. Helvetian Press. ISBN 978-0956478115. OCLC 760830351.
- ^ Currell BR, van Dam-Mieras MC (1997). Biotechnological Innovations in Chemical Synthesis. Oxford: Butterworth-Heinemann. pp. 125–128. ISBN 978-0-7506-0561-8.
- ^ Abrusan G, Marsh JA (2019). "Ligand Binding Site Structure Shapes Folding, Assembly and Degradation of Homomeric Protein Complexes". Journal of Molecular Biology. 431 (19): 3871–3888. doi:10.1016/j.jmb.2019.07.014. PMC 6739599. PMID 31306664.
- ^ Abrusan G, Marsh JA (2018). "Ligand Binding Site Structure Influences the Evolution of Protein Complex Function and Topology". Cell Reports. 22 (12): 3265–3276. doi:10.1016/j.celrep.2018.02.085. PMC 5873459. PMID 29562182.
- ^ Abrusan G, Marsh JA (2019). "Ligand-Binding-Site Structure Shapes Allosteric Signal Transduction and the Evolution of Allostery in Protein Complexes". Molecular Biology and Evolution. 36 (8): 1711–1727. doi:10.1093/molbev/msz093. PMC 6657754. PMID 31004156.
- ^ a b Cimermancic P, Weinkam P, Rettenmaier TJ, Bichmann L, Keedy DA, Woldeyes RA, et al. (February 2016). "CryptoSite: Expanding the Druggable Proteome by Characterization and Prediction of Cryptic Binding Sites". Journal of Molecular Biology. 428 (4): 709–719. doi:10.1016/j.jmb.2016.01.029. PMID 26854760.
- ^ Beglov D, Hall DR, Wakefield AE, Luo L, Allen KN, Kozakov D, et al. (April 2018). "Exploring the structural origins of cryptic sites on proteins". Proceedings of the National Academy of Sciences of the United States of America. 115 (15): E3416–E3425. doi:10.1073/pnas.1711490115. PMID 29581267.
- ^ Bowman GR, Bolin ER, Hart KM, Maguire BC, Marqusee S (March 2015). "Discovery of multiple hidden allosteric sites by combining Markov state models and experiments". Proceedings of the National Academy of Sciences of the United States of America. 112 (9): 2734–9. doi:10.1073/pnas.1417811112. PMC 4352775. PMID 25730859.
- ^ Iida S, Nakamura HK, Mashimo T, Fukunishi Y (November 2020). "Structural Fluctuations of Aromatic Residues in an Apo-Form Reveal Cryptic Binding Sites: Implications for Fragment-Based Drug Design". The Journal of Physical Chemistry. B. 124 (45): 9977–9986. doi:10.1021/acs.jpcb.0c04963. PMID 33140952.
- ^ Ahern K (January 2017). "Teaching biochemistry online at Oregon State University". Biochemistry and Molecular Biology Education. 45 (1): 25–30. doi:10.1002/bmb.20979. PMID 27228905.
- ^ Anne A, Demaille C (October 2012). "Kinetics of enzyme action on surface-attached substrates: a practical guide to progress curve analysis in any kinetic situation". Langmuir. 28 (41): 14665–71. doi:10.1021/la3030827. PMID 22978617.
- ^ Morris JR, Hartl DL, Knoll AH. Biology: how life works (Second ed.). New York, NY. ISBN 9781464126093. OCLC 937824456.
- ^ Spitzer R, Cleves AE, Varela R, Jain AN (April 2014). "Protein function annotation by local binding site surface similarity". Proteins. 82 (4): 679–94. doi:10.1002/prot.24450. PMC 3949165. PMID 24166661.
- ^ Peng J, Li XP (November 2018). "Apolipoprotein A-IV: A potential therapeutic target for atherosclerosis". Prostaglandins & Other Lipid Mediators. 139: 87–92. doi:10.1016/j.prostaglandins.2018.10.004. PMID 30352313.
- ^ McNamara JW, Sadayappan S (December 2018). "Skeletal myosin binding protein-C: An increasingly important regulator of striated muscle physiology". Archives of Biochemistry and Biophysics. 660: 121–128. doi:10.1016/j.abb.2018.10.007. PMC 6289839. PMID 30339776.
- ^ a b Widemann BC, Adamson PC (June 2006). "Understanding and managing methotrexate nephrotoxicity". The Oncologist. 11 (6): 694–703. doi:10.1634/theoncologist.11-6-694. PMID 16794248.
- ^ a b Rajagopalan PT, Zhang Z, McCourt L, Dwyer M, Benkovic SJ, Hammes GG (October 2002). "Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics". Proceedings of the National Academy of Sciences of the United States of America. 99 (21): 13481–6. doi:10.1073/pnas.172501499. PMC 129699. PMID 12359872.
- ^ Frishman WH, Cheng-Lai A, Chen J, eds. (2000). Current Cardiovascular Drugs. doi:10.1007/978-1-4615-6767-7. ISBN 978-1-57340-135-7.
- ^ Montecucco C, Molgó J (June 2005). "Botulinal neurotoxins: revival of an old killer". Current Opinion in Pharmacology. 5 (3): 274–9. doi:10.1016/j.coph.2004.12.006. PMID 15907915.
- ^ Roche DB, Brackenridge DA, McGuffin LJ (December 2015). "Proteins and Their Interacting Partners: An Introduction to Protein-Ligand Binding Site Prediction Methods". International Journal of Molecular Sciences. 16 (12): 29829–42. doi:10.3390/ijms161226202. PMC 4691145. PMID 26694353.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Broomhead NK, Soliman ME (March 2017). "Can We Rely on Computational Predictions To Correctly Identify Ligand Binding Sites on Novel Protein Drug Targets? Assessment of Binding Site Prediction Methods and a Protocol for Validation of Predicted Binding Sites". Cell Biochemistry and Biophysics. 75 (1): 15–23. doi:10.1007/s12013-016-0769-y. PMID 27796788.
- ^ Jones S, Thornton JM (September 1997). "Analysis of protein-protein interaction sites using surface patches". Journal of Molecular Biology. 272 (1): 121–32. doi:10.1006/jmbi.1997.1234. PMID 9299342.
External links
- Binding Sites at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Drawing the active site of an enzyme
