P4HB
Protein disulfide-isomerase, also known as the beta-subunit of prolyl 4-hydroxylase (P4HB), is an enzyme that in humans encoded by the P4HB gene. The human P4HB gene is localized in chromosome 17q25.[5][6][7][8] Unlike other prolyl 4-hydroxylase family proteins, this protein is multifunctional and acts as an oxidoreductase for disulfide formation, breakage, and isomerization.[9] The activity of P4HB is tightly regulated. Both dimer dissociation and substrate binding are likely to enhance its enzymatic activity during the catalysis process.[10][11]
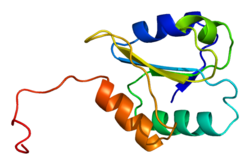
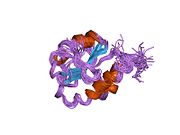
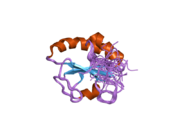
Structure
P4HB has four thioredoxin domains (a, b, b’, and a’), with two CGHC active sites in the a and a’ domains. In both the reduced and oxidized state, these domains are arranged as a horseshoe shape. In reduced P4HB, domains a, b, and b' are in the same plane, while domain a' twists out at an ∼45° angle. When oxidized, the four domains stay in the same plane, and the distance between the active sites is larger than that in the reduced state. The oxidized form also exposes more hydrophobic areas and possesses a larger cleft to facilitate substrate binding.[12][13] P4HB has been shown to dimerize in vivo via noncatalytic bb' domains. Formation of dimer blocks substrate-binding site and inhibits P4HB’s activity.[14]
Function
This gene encodes the beta subunit of prolyl 4-hydroxylase, a highly abundant multifunctional enzyme that belongs to the protein disulfide isomerase family. When present as a tetramer consisting of two alpha and two beta subunits, this enzyme is involved in hydroxylation of prolyl residues in preprocollagen. This enzyme is also a disulfide isomerase containing two thioredoxin domains that catalyze the formation, breakage and rearrangement of disulfide bonds. Other known functions include its ability to act as a chaperone that inhibits aggregation of misfolded proteins in a concentration-dependent manner, its ability to bind thyroid hormone, its role in both the influx and efflux of S-nitrosothiol-bound nitric oxide, and its function as a subunit of the microsomal triglyceride transfer protein complex.[6]
Clinical significance
P4HB is a risk factor of amyotrophic lateral sclerosis (ALS). Studies have shown significant genotypic associations for two SNPs in P4HB with FALS (familial ALS), rs876016 (P=0.0198) and rs2070872 (P=0.0046). Together with other ER stress markers, P4HB is greatly elevated in ALS spinal cord.[15] P4HB can also be nitrosylated, and elevation of nitrosylated P4HB has been shown in Parkinson's and Alzheimer's disease brain tissue, as well as in transgenic mutant superoxide dismutase 1 mouse and human sporadic amyotrophic lateral sclerosis spinal cord tissues.[16][17] In addition to neurodegenerative diseases, P4HB level is upregulated in glioblastoma multiforme (GBM) (brain tumor). Inhibition of P4HB attenuates resistance to temozolomide, a standard GBM chemotherapeutic agent, via the PERK arm of endoplasmic reticulum stress response pathway.[18] Furthermore, heterozygous missense mutation in P4HB can cause Cole-Carpenter syndrome, a severe bone fragility disorder.[19]
Interactions
P4HB has been shown to interact with UBQLN1,[20] ERO1LB[21][22] and ERO1L.[21][22]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000185624 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000025130 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Shoulders CC, Brett DJ, Bayliss JD, Narcisi TM, Jarmuz A, Grantham TT, Leoni PR, Bhattacharya S, Pease RJ, Cullen PM (December 1993). "Abetalipoproteinemia is caused by defects of the gene encoding the 97 kDa subunit of a microsomal triglyceride transfer protein". Human Molecular Genetics. 2 (12): 2109–16. doi:10.1093/hmg/2.12.2109. PMID 8111381.
- ^ a b "Entrez Gene: P4HB procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide".
- ^ Galligan JJ, Petersen DR (July 2012). "The human protein disulfide isomerase gene family". Human Genomics. 6 (6): 6. doi:10.1186/1479-7364-6-6. PMC 3500226. PMID 23245351.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Pajunen L, Jones TA, Goddard A, Sheer D, Solomon E, Pihlajaniemi T, Kivirikko KI (1991-01-01). "Regional assignment of the human gene coding for a multifunctional polypeptide (P4HB) acting as the beta-subunit of prolyl 4-hydroxylase and the enzyme protein disulfide isomerase to 17q25". Cytogenetics and Cell Genetics. 56 (3–4): 165–8. PMID 1647289.
- ^ Lumb RA, Bulleid NJ (December 2002). "Is protein disulfide isomerase a redox-dependent molecular chaperone?". The EMBO Journal. 21 (24): 6763–70. PMC 139105. PMID 12485997.
- ^ Bastos-Aristizabal S, Kozlov G, Gehring K (May 2014). "Structural insight into the dimerization of human protein disulfide isomerase". Protein Science. 23 (5): 618–26. doi:10.1002/pro.2444. PMC 4005713. PMID 24549644.
- ^ Winter J, Klappa P, Freedman RB, Lilie H, Rudolph R (January 2002). "Catalytic activity and chaperone function of human protein-disulfide isomerase are required for the efficient refolding of proinsulin". The Journal of Biological Chemistry. 277 (1): 310–7. doi:10.1074/jbc.M107832200. PMID 11694508.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H (January 2006). "The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites". Cell. 124 (1): 61–73. doi:10.1016/j.cell.2005.10.044. PMID 16413482.
- ^ Wang C, Li W, Ren J, Fang J, Ke H, Gong W, Feng W, Wang CC (July 2013). "Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase". Antioxidants & Redox Signaling. 19 (1): 36–45. doi:10.1089/ars.2012.4630. PMID 22657537.
- ^ Bastos-Aristizabal S, Kozlov G, Gehring K (May 2014). "Structural insight into the dimerization of human protein disulfide isomerase". Protein Science. 23 (5): 618–26. doi:10.1002/pro.2444. PMC 4005713. PMID 24549644.
- ^ Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK (June 2008). "Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis". Neurobiology of Disease. 30 (3): 400–7. doi:10.1016/j.nbd.2008.02.009. PMID 18440237.
- ^ Walker AK, Farg MA, Bye CR, McLean CA, Horne MK, Atkin JD (January 2010). "Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis". Brain. 133 (Pt 1): 105–16. doi:10.1093/brain/awp267. PMID 19903735.
- ^ Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA (May 2006). "S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration". Nature. 441 (7092): 513–7. doi:10.1038/nature04782. PMID 16724068.
- ^ Sun S, Lee D, Ho AS, Pu JK, Zhang XQ, Lee NP, Day PJ, Lui WM, Fung CF, Leung GK (May 2013). "Inhibition of prolyl 4-hydroxylase, beta polypeptide (P4HB) attenuates temozolomide resistance in malignant glioma via the endoplasmic reticulum stress response (ERSR) pathways". Neuro-Oncology. 15 (5): 562–77. doi:10.1093/neuonc/not005. PMC 3635523. PMID 23444257.
- ^ Rauch F, Fahiminiya S, Majewski J, Carrot-Zhang J, Boudko S, Glorieux F, Mort JS, Bächinger HP, Moffatt P (March 2015). "Cole-Carpenter syndrome is caused by a heterozygous missense mutation in P4HB". American Journal of Human Genetics. 96 (3): 425–31. doi:10.1016/j.ajhg.2014.12.027. PMC 4375435. PMID 25683117.
- ^ Ko HS, Uehara T, Nomura Y (September 2002). "Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death". The Journal of Biological Chemistry. 277 (38): 35386–92. doi:10.1074/jbc.M203412200. PMID 12095988.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R (February 2002). "ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family". The EMBO Journal. 21 (4): 835–44. doi:10.1093/emboj/21.4.835. PMC 125352. PMID 11847130.
- ^ a b Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R (November 2001). "Manipulation of oxidative protein folding and PDI redox state in mammalian cells". The EMBO Journal. 20 (22): 6288–96. doi:10.1093/emboj/20.22.6288. PMC 125306. PMID 11707400.
Further reading
- Galligan JJ, Petersen DR (July 2012). "The human protein disulfide isomerase gene family". Human Genomics. 6 (6): 6. doi:10.1186/1479-7364-6-6. PMC 3500226. PMID 23245351.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Wilkinson B, Gilbert HF (June 2004). "Protein disulfide isomerase". Biochimica et Biophysica Acta. 1699 (1–2): 35–44. doi:10.1016/j.bbapap.2004.02.017. PMID 15158710.
- Pihlajaniemi T, Myllylä R, Kivirikko KI (1992). "Prolyl 4-hydroxylase and its role in collagen synthesis". Journal of Hepatology. 13 Suppl 3: S2-7. doi:10.1016/0168-8278(91)90002-S. PMID 1667665.
- Wilkinson B, Gilbert HF (June 2004). "Protein disulfide isomerase". Biochimica et Biophysica Acta. 1699 (1–2): 35–44. doi:10.1016/j.bbapap.2004.02.017. PMID 15158710.
- Hochstrasser DF, Frutiger S, Paquet N, Bairoch A, Ravier F, Pasquali C, Sanchez JC, Tissot JD, Bjellqvist B, Vargas R (December 1992). "Human liver protein map: a reference database established by microsequencing and gel comparison". Electrophoresis. 13 (12): 992–1001. doi:10.1002/elps.11501301201. PMID 1286669.
- Chessler SD, Byers PH (April 1992). "Defective folding and stable association with protein disulfide isomerase/prolyl hydroxylase of type I procollagen with a deletion in the pro alpha 2(I) chain that preserves the Gly-X-Y repeat pattern". The Journal of Biological Chemistry. 267 (11): 7751–7. PMID 1339453.
- Vuori K, Myllylä R, Pihlajaniemi T, Kivirikko KI (April 1992). "Expression and site-directed mutagenesis of human protein disulfide isomerase in Escherichia coli. This multifunctional polypeptide has two independently acting catalytic sites for the isomerase activity". The Journal of Biological Chemistry. 267 (11): 7211–4. PMID 1559965.
- Tasanen K, Oikarinen J, Kivirikko KI, Pihlajaniemi T (June 1992). "Promoter of the gene for the multifunctional protein disulfide isomerase polypeptide. Functional significance of the six CCAAT boxes and other promoter elements". The Journal of Biological Chemistry. 267 (16): 11513–9. PMID 1597478.
- Bauw G, Rasmussen HH, van den Bulcke M, van Damme J, Puype M, Gesser B, Celis JE, Vandekerckhove J (July 1990). "Two-dimensional gel electrophoresis, protein electroblotting and microsequencing: a direct link between proteins and genes". Electrophoresis. 11 (7): 528–36. doi:10.1002/elps.1150110703. PMID 1699755.
- Ward LD, Hong J, Whitehead RH, Simpson RJ (October 1990). "Development of a database of amino acid sequences for human colon carcinoma proteins separated by two-dimensional polyacrylamide gel electrophoresis". Electrophoresis. 11 (10): 883–91. doi:10.1002/elps.1150111019. PMID 2079031.
- Tasanen K, Parkkonen T, Chow LT, Kivirikko KI, Pihlajaniemi T (November 1988). "Characterization of the human gene for a polypeptide that acts both as the beta subunit of prolyl 4-hydroxylase and as protein disulfide isomerase". The Journal of Biological Chemistry. 263 (31): 16218–24. PMID 2846539.
- Koivu J, Myllylä R, Helaakoski T, Pihlajaniemi T, Tasanen K, Kivirikko KI (May 1987). "A single polypeptide acts both as the beta subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase". The Journal of Biological Chemistry. 262 (14): 6447–9. PMID 3032969.
- Pihlajaniemi T, Helaakoski T, Tasanen K, Myllylä R, Huhtala ML, Koivu J, Kivirikko KI (March 1987). "Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene". The EMBO Journal. 6 (3): 643–9. PMC 553446. PMID 3034602.
- Morris JI, Varandani PT (February 1988). "Characterization of a cDNA for human glutathione-insulin transhydrogenase (protein-disulfide isomerase/oxidoreductase)". Biochimica et Biophysica Acta. 949 (2): 169–80. doi:10.1016/0167-4781(88)90080-2. PMID 3342239.
- Gosden JR, Middleton PG, Rout D, De Angelis C (1987). "Chromosomal localization of the human oncogene ERBA2". Cytogenetics and Cell Genetics. 43 (3–4): 150–3. doi:10.1159/000132313. PMID 3467900.
- Cheng SY, Gong QH, Parkison C, Robinson EA, Appella E, Merlino GT, Pastan I (August 1987). "The nucleotide sequence of a human cellular thyroid hormone binding protein present in endoplasmic reticulum". The Journal of Biological Chemistry. 262 (23): 11221–7. PMID 3611107.
- Helaakoski T, Annunen P, Vuori K, MacNeil IA, Pihlajaniemi T, Kivirikko KI (May 1995). "Cloning, baculovirus expression, and characterization of a second mouse prolyl 4-hydroxylase alpha-subunit isoform: formation of an alpha 2 beta 2 tetramer with the protein disulfide-isomerase/beta subunit". Proceedings of the National Academy of Sciences of the United States of America. 92 (10): 4427–31. doi:10.1073/pnas.92.10.4427. PMC 41957. PMID 7753822.
- Kemmink J, Darby NJ, Dijkstra K, Scheek RM, Creighton TE (December 1995). "Nuclear magnetic resonance characterization of the N-terminal thioredoxin-like domain of protein disulfide isomerase". Protein Science. 4 (12): 2587–93. doi:10.1002/pro.5560041216. PMC 2143042. PMID 8580850.
- Kemmink J, Darby NJ, Dijkstra K, Nilges M, Creighton TE (June 1996). "Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy". Biochemistry. 35 (24): 7684–91. doi:10.1021/bi960335m. PMID 8672469.
- Ji H, Reid GE, Moritz RL, Eddes JS, Burgess AW, Simpson RJ (1997). "A two-dimensional gel database of human colon carcinoma proteins". Electrophoresis. 18 (3–4): 605–13. doi:10.1002/elps.1150180344. PMID 9150948.