Eyespot apparatus

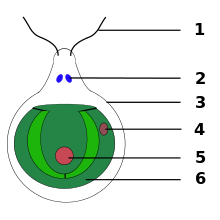
The eyespot apparatus (or stigma) is a photoreceptive organelle found in the flagellate or (motile) cells of green algae and other unicellular photosynthetic organisms such as euglenids. It allows the cells to sense light direction and intensity and respond to it, prompting the organism to either swim towards the light (positive phototaxis), or away from it (negative phototaxis). A related response ("photoshock" or photophobic response) occurs when cells are briefly exposed to high light intensity, causing the cell to stop, briefly swim backwards, then change swimming direction. Eyespot-mediated light perception helps the cells in finding an environment with optimal light conditions for photosynthesis. Eyespots are the simplest and most common "eyes" found in nature, composed of photoreceptors and areas of bright orange-red red pigment granules.[1] Signals relayed from the eyespot photoreceptors result in alteration of the beating pattern of the flagella, generating a phototactic response.[2]
Microscopic structure
[edit]Under the light microscope, eyespots appear as dark, orange-reddish spots or stigmata. They get their color from carotenoid pigments contained in bodies called pigment granules. The photoreceptors are found in the plasma membrane overlaying the pigmented bodies.
The eyespot apparatus of Euglena comprises the paraflagellar body connecting the eyespot to the flagellum. In electron microscopy, the eyespot apparatus appears as a highly ordered lamellar structure formed by membranous rods in a helical arrangement.[3]
In Chlamydomonas, the eyespot is part of the chloroplast and takes on the appearance of a membranous sandwich structure. It is assembled from chloroplast membranes (outer, inner, and thylakoid membranes) and carotenoid-filled granules overlaid by plasma membrane. The stacks of granules act as a quarter-wave plate, reflecting incoming photons back to the overlying photoreceptors, while shielding the photoreceptors from light coming from other directions. It disassembles during cell division and reforms in the daughter cells in an asymmetric fashion in relation to the cytoskeleton. This asymmetric positioning of the eyespot in the cell is essential for proper phototaxis.[4]
Eyespot proteins
[edit]The most critical eyespot proteins are the photoreceptor proteins that sense light. The photoreceptors found in unicellular organisms fall into two main groups: flavoproteins and retinylidene proteins (rhodopsins). Flavoproteins are characterized by containing flavin molecules as chromophores, whereas retinylidene proteins contain retinal. The photoreceptor protein in Euglena is likely a flavoprotein.[3] In contrast, Chlamydomonas phototaxis is mediated by archaeal-type rhodopsins.[5]
Besides photoreceptor proteins, eyespots contain a large number of structural, metabolic and signaling proteins. The eyespot proteome of Chlamydomonas cells consists of roughly 200 different proteins.[6]
Photoreception and signal transduction
[edit]The Euglena photoreceptor was identified as a blue-light-activated adenylyl cyclase.[7] Excitation of this receptor protein results in the formation of cyclic adenosine monophosphate (cAMP) as a second messenger. Chemical signal transduction ultimately triggers changes in flagellar beat patterns and cell movement.
The archaeal-type rhodopsins of Chlamydomonas contain an all-trans retinylidene chromatophore which undergoes photoisomerization to a 13-cis isomer. This activates a photoreceptor channel, leading to a change in membrane potential and cellular calcium ion concentration.[5] Photoelectric signal transduction ultimately triggers changes in flagellar strokes and thus cell movement.[2]
See also
[edit]References
[edit]- ^ Kreimer, G. (2009). "The green algal eyespot apparatus: A primordial visual system and more?". Current Genetics. 55 (1): 19–43. doi:10.1007/s00294-008-0224-8. PMID 19107486. S2CID 8011518.
- ^ a b Hegemann P (1997). "Vision in microalgae". Planta. 203 (3): 265–74. Bibcode:1997Plant.203..265H. doi:10.1007/s004250050191. PMID 9431675. S2CID 11933925.
- ^ a b Wolken J (1977). "Euglena: the photoreceptor system for phototaxis". J Protozool. 24 (4): 518–22. doi:10.1111/j.1550-7408.1977.tb01004.x. PMID 413913.
- ^ Dieckmann C (2003). "Eyespot placement and assembly in the green alga Chlamydomonas". BioEssays. 25 (4): 410–6. doi:10.1002/bies.10259. PMID 12655648.
- ^ a b Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, Tabata S, Miura K, Fukuzawa H, Nakamura S, Takahashi T (2003). "Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization". Biochem Biophys Res Commun. 301 (3): 711–7. doi:10.1016/S0006-291X(02)03079-6. PMID 12565839.
- ^ Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, Fiedler M, Mittag M, Kreimer G (2006). "Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements". Plant Cell. 18 (8): 1908–30. doi:10.1105/tpc.106.041749. PMC 1533972. PMID 16798888.
- ^ Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M (2002). "A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis". Nature. 415 (6875): 1047–51. Bibcode:2002Natur.415.1047I. doi:10.1038/4151047a. PMID 11875575. S2CID 4420996.
