User:Lavenderviolet/sandbox
RNA Immunoprecipitation chip
[edit]RIP-chip is a molecular biology technique which combines RNA immunoprecipitation with a microarray. The purpose of this technique is to identify which RNA sequences interact with a particular RNA binding protein of interest in vivo[1][2][3][4]. It can also be used to determine relative levels of gene expression, to identify subsets of RNAs which may be co-regulated, or to identify RNAs that may have related functions[4][5]. This technique provides insight into the post-transcriptional gene regulation which occurs between RNA and RNA binding proteins[5].
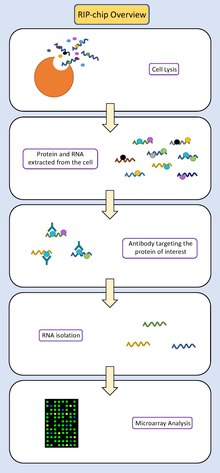
- Collect and lyse the cells of interest.
- Isolate all RNA fragments and the proteins bound to them from the solution.
- Immunoprecipitate the protein of interest. The solution containing the protein bound RNAs is washed over beads which have been conjugated to antibodies. These antibodies are designed to bind to the protein of interest. They pull the protein (and any RNA fragments that are specifically bound to it) out of the solution which contains the rest of the cell contents.
- Dissociate the protein bound RNA from the antibody-bead complex. Then, use a centrifuge to separate the protein-bound RNA from the heavier antibody-bead complexes, keeping the protein bound RNA and discarding the beads.
- Disassociate the RNA from the protein of interest.
- Isolate the RNA fragments from the protein using a centrifuge.
- Use Reverse Transcription PCR to convert the RNA fragments into cDNA (DNA that is complementary to the RNA fragments).
- Fluorescently label these cDNA fragments.
- Prepare the gene chip. This is a small chip that has DNA sequences bound to it in known locations. These DNA sequences correspond to all of the known genes in the genome of the organism that the researcher is working with (or a subset of genes that the researcher is interested in). The cDNA sequences that have been collected will be complementary to some of these DNA sequences, as the cDNAs represent a subset of the RNAs transcribed from the genome.
- Allow the cDNA fragments to competitively hybridize to the DNA sequences bound to the chip.
- Detection of the fluorescent signal from the cDNA bound to the chip tells researchers which gene(s) on the chip were hybridized to the cDNA.
The genes fluorescently identified by the chip analysis are the genes whose RNA interacts with the original protein of interest. The strength of the fluorescent signal for a particular gene can indicate how much of that particular RNA was present in the original sample, which indicates the expression level of that gene.
Development and Similar Techniques
[edit]Previous techniques aiming to understand protein-RNA interactions included RNA Electrophoretic Mobility Shift Assays and UV-crosslinking[9], however these techniques cannot be used when the RNA sequence is unknown[1]. To resolve this, RIP-chip combines RNA immunoprecipitation to isolate RNA molecules interacting with specific proteins with a microarray which can elucidate the identity of the RNAs participating in this interaction[1][5]. Alternatives to RIP-chip include:
- RIP-seq: Involves sequencing the RNAs that were pulled down using high-throughput sequencing rather than analyzing them with a microarray[5]. Authors Zhao et al., 2010[10] combined the RNA immunoprecipitation procedure with RNA sequencing. Using specific antibodies (α-Ezh2) they immunoprecipitated nuclear RNA isolated from mouse ES cells, and subsequently sequenced the pulled-down RNA using the next generation sequencing platform, Illumina[10].
- CLIP-seq: The RNA binding protein is cross-linked to the RNA via the use of UV light prior to immunoprecipitation[11]. Authors Licatalosi et al., 2008[12] first combined the UV crosslinking coupled immunoprecipitation procedure (CLIP) with high throughput sequencing methods to determine Nova-RNA binding sites in the mouse brain[11][12]. In addition, they determined that this protocol could determine de novo protein interactions[12].
- ChIP-on-chip: A similar technique which detects the binding of proteins to genomic DNA rather than RNA[13].
References
[edit]- ^ a b c Gagliardi, Miriam; Matarazzo, Maria R. (2016), Lanzuolo, Chiara; Bodega, Beatrice (eds.), "RIP: RNA Immunoprecipitation", Polycomb Group Proteins, vol. 1480, New York, NY: Springer New York, pp. 73–86, doi:10.1007/978-1-4939-6380-5_7, ISBN 978-1-4939-6378-2, retrieved 2020-10-24
- ^ Townley-Tilson, W.H. D. (2006-09-06). "Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein". RNA. 12 (10): 1853–1867. doi:10.1261/rna.76006. ISSN 1355-8382. PMC 1581977. PMID 16931877.
{{cite journal}}: CS1 maint: PMC format (link) - ^ Khalil, Ahmad M.; Guttman, Mitchell; Huarte, Maite; Garber, Manuel; Raj, Arjun; Rivea Morales, Dianali; Thomas, Kelly; Presser, Aviva; Bernstein, Bradley E.; van Oudenaarden, Alexander; Regev, Aviv (2009-07-14). "Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression". Proceedings of the National Academy of Sciences. 106 (28): 11667–11672. doi:10.1073/pnas.0904715106. ISSN 0027-8424. PMC 2704857. PMID 19571010.
{{cite journal}}: CS1 maint: PMC format (link) - ^ a b Hendrickson, David G.; Hogan, Daniel J.; Herschlag, Daniel; Ferrell, James E.; Brown, Patrick O. (2008-05-07). Bähler, Jürg (ed.). "Systematic Identification of mRNAs Recruited to Argonaute 2 by Specific microRNAs and Corresponding Changes in Transcript Abundance". PLoS ONE. 3 (5): e2126. doi:10.1371/journal.pone.0002126. ISSN 1932-6203. PMC 2330160. PMID 18461144.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ a b c d Jayaseelan, Sabarinath; Doyle, Francis; Tenenbaum, Scott A. (2014-05). "Profiling post-transcriptionally networked mRNA subsets using RIP-Chip and RIP-Seq". Methods. 67 (1): 13–19. doi:10.1016/j.ymeth.2013.11.001. PMC 4004666. PMID 24257445.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Baroni, Timothy E.; Chittur, Sridar V.; George, Ajish D.; Tenenbaum, Scott A. (2008), Wilusz, Jeffrey (ed.), "Advances in RIP-Chip Analysis: RNA-Binding Protein Immunoprecipitation-Microarray Profiling", Post-Transcriptional Gene Regulation, Methods In Molecular Biology™, Totowa, NJ: Humana Press, pp. 93–108, doi:10.1007/978-1-59745-033-1_6#fig6_1_978-1-59745-033-1, ISBN 978-1-59745-033-1, retrieved 2020-10-25
- ^ Conrad, Nicholas K. (2008-01-01), Maquat, Lynne E.; Kiledjian, Megerditch (eds.), "Chapter 15 - Co-Immunoprecipitation Techniques for Assessing RNA–Protein Interactions In Vivo", Methods in Enzymology, RNA Turnover in Eukaryotes: Analysis of Specialized and Quality Control RNA Decay Pathways, vol. 449, Academic Press, pp. 317–342, retrieved 2020-10-25
- ^ Niranjanakumari, Somashe; Lasda, Erika; Brazas, Robert; Garcia-Blanco, Mariano A. (2002-02-01). "Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo". Methods. 26 (2): 182–190. doi:10.1016/S1046-2023(02)00021-X. ISSN 1046-2023.
- ^ Thomson, A.M.; Rogers, J.T.; Walker, C.E.; Staton, J.M.; Leedman, P.J. (1999-11). "Optimized RNA Gel-Shift and UV Cross-Linking Assays for Characterization of Cytoplasmic RNA-Protein Interactions". BioTechniques. 27 (5): 1032–1042. doi:10.2144/99275rr03. ISSN 0736-6205.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Zhao, Jing; Ohsumi, Toshiro K.; Kung, Johnny T.; Ogawa, Yuya; Grau, Daniel J.; Sarma, Kavitha; Song, Ji Joon; Kingston, Robert E.; Borowsky, Mark; Lee, Jeannie T. (2010-12). "Genome-wide Identification of Polycomb-Associated RNAs by RIP-seq". Molecular Cell. 40 (6): 939–953. doi:10.1016/j.molcel.2010.12.011. PMC 3021903. PMID 21172659.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ a b Milek, Miha; Wyler, Emanuel; Landthaler, Markus (2012-04). "Transcriptome-wide analysis of protein–RNA interactions using high-throughput sequencing". Seminars in Cell & Developmental Biology. 23 (2): 206–212. doi:10.1016/j.semcdb.2011.12.001.
{{cite journal}}: Check date values in:|date=(help) - ^ a b c Licatalosi, Donny D.; Mele, Aldo; Fak, John J.; Ule, Jernej; Kayikci, Melis; Chi, Sung Wook; Clark, Tyson A.; Schweitzer, Anthony C.; Blume, John E.; Wang, Xuning; Darnell, Jennifer C. (2008-11). "HITS-CLIP yields genome-wide insights into brain alternative RNA processing". Nature. 456 (7221): 464–469. doi:10.1038/nature07488. ISSN 0028-0836. PMC 2597294. PMID 18978773.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: PMC format (link) - ^ Ansorge, Wilhelm J. (2010), "Novel Next-Generation DNA Sequencing Techniques for Ultra High-Throughput Applications in Bio-Medicine", Molecular Diagnostics, Elsevier, pp. 365–378, doi:10.1016/b978-0-12-374537-8.00024-9, ISBN 978-0-12-374537-8, retrieved 2020-10-24

