User:Vrabiochemhw
| Isocitrate Lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Homotetrameric structure of Isocitrate lyase from E. coli. From PDB 1IGW[1]. | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.3.1 | ||||||||
| CAS no. | 9045-78-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Isocitrate lyase (EC 4.1.3.1), or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate [2][3][4]. Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle (TCA cycle) and is used by bacteria, fungi, and plants [4].
The systematic name of this enzyme class is isocitrate glyoxylate-lyase (succinate-forming). Other names in common use include isocitrase, isocitritase, isocitratase, threo-Ds-isocitrate glyoxylate-lyase, and isocitrate glyoxylate-lyase. This enzyme participates in glyoxylate and dicarboxylate metabolism.
Mechanism
[edit]This enzyme belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. Other enzymes also belong to this family including carboxyvinyl-carboxyphosphonate phosphorylmutase (EC 2.7.8.23) which catalyses the conversion of 1-carboxyvinyl carboxyphosphonate to 3-(hydrohydroxyphosphoryl) pyruvate carbon dioxide, and phosphoenolpyruvate mutase (EC 5.4.2.9), which is involved in the biosynthesis of phosphinothricin tripeptide antibiotics.
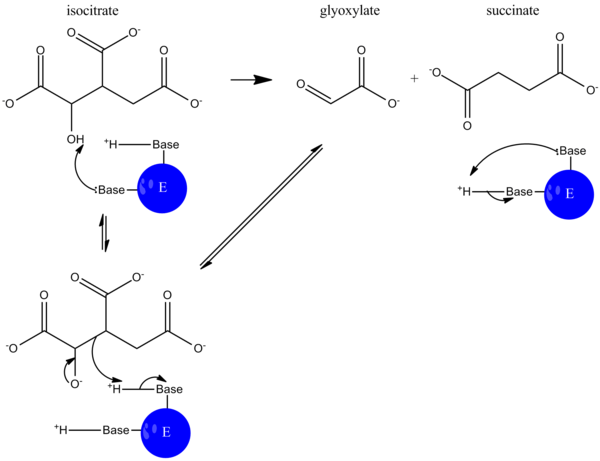
During catalysis, isocitrate is deprotonated, and an aldol cleavage results in the release of succinate and glyoxylate. This reaction mechanism functions much like that of aldolase in glycolysis[5].
In the glyoxylate cycle, malate synthase then catalyzes the condensation of glyoxylate and acetyl-CoA to form malate so the cycle can continue.
ICL competes with isocitrate dehydrogenase, an enzyme found in the TCA cycle, for isocitrate processing. Flux through these enzymes is controlled by phosphorylation of isocitrate dehydrogenase, which has a much higher affinity compared to ICL for isocitrate[6].
Enzyme Structure
[edit]As of late 2007, 5 structures have been solved for this class of enzymes, with PDB accession codes 1DQU, 1F61, 1F8I, 1F8M, and 1IGW.
ICL is composed of four identical chains and requires a Mg2+ or Mn2+ and a thiol for activity[4]. In Escherichia coli, Lys-193, Lys-194, Cys-195, His-197, and His-356 are thought to be catalytic residues, while His-184 is thought to be involved in the assembly of the tetrameric enzyme[7].
Between prokaryotes and eukaryotes, a difference in ICL structure is the addition of approximately 100 amino acids near the center of the eukaryotic enzyme. These are thought to function in the localization of ICL to single-membrane-bound organelles called glyoxysomes[4] [8] . These additional amino acids account for the difference in molecular mass: the prokaryotic ICL is 48kDa, while the eukaryotic ICL is 67 kDa[4] . Only one cysteine residue is conserved between the sequences of the fungal, plant and bacterial enzymes; it is located in the middle of a conserved hexapeptide.
Biological Function
[edit]The ICL enzyme has been found to be functional in various archaea, bacteria, protists, plants, fungi, and nematodes[9]. Although the gene has been found in genomes of nematodes and cnidaria, it has not been found in the genomes of placental mammals[9].
By diverting isocitrate from the TCA cycle, the actions of ICL and malate synthase in the glyoxylate cycle result in the net assimilation of carbon from 2-carbon compounds[10] . Thus, while the TCA cycle yields no net carbon assimilation, the glyoxylate cycle generates intermediates that can be used to synthesize glucose (via gluconeogenesis), and other biosynthetic products. As a result, organisms that use ICL and malate synthase are able to synthesize glucose and TCA intermediates from acetyl-CoA derived from acetate or from the degradation of ethanol, fatty acids or poly-β-hydroxybutyrate[4].
This function is especially important for higher plants which use oilseeds. In these germinating seeds, the breakdown of oils generates acetyl-CoA. This serves as a substrate for the glyoxylate cycle, which generates other cyclic intermediates and serves as a primary nutrient source prior to the production of sugars from photosynthesis[8].
Disease Relevance
[edit]ICL has found to be important in human, animal, and plant pathogenesis[4]. For several agricultural crops including cereals, cucumbers, and melons, increased expression of the gene encoding ICL is important for fungal virulence[4] . For instance, increased gene expression of icl1 has been seen in the fungus Leptosphaeria maculans upon infection of canola. Inactivation of the icl1 gene leads to reduced pathogenicity of the fungus, which is thought to be a result of the inability of the fungus to use carbon sources provided by the plant[11] .
Additionally, upregulation of the glyoxylate cycle has been seen for pathogens that attack humans. This is the case for fungi such as Candida albicans, which inhabits the skin, mouth, GI tract, gut and vagina of mammals and can lead to systemic infections of immunocompromised patients; as well as for the bacterium Mycobacterium tuberculosis, the major causative agent of tuberculosis[12][13]. In this latter case, ICL has been found to be essential for survival in the host[14].
See also
[edit]References
[edit]- ^ Britton, K. L.; Abeysinghe, I. S.; Baker, P. J.; Barynin, V.; Diehl, P.; Langridge, S. J.; McFadden, B. A.; Sedelnikova, S. E.; Stillman, T. J.; Weeradechapon, K.; Rice, D. W. (2001). "The structure and domain organization of Escherichia coli isocitrate lyase". Acta Crystallogr D Biol Crystallogr. 57 (9): 1209–1218. doi:10.1107/s0907444901008642. PMID 11526312.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) - ^ Beeching JR (1989). "High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis". Protein Seq. Data Anal. 2 (6): 463–466. PMID 2696959.
- ^ Tanaka A, Atomi H, Ueda M, Hikida M, Hishida T, Teranishi Y (1990). "Peroxisomal isocitrate lyase of the n-alkane-assimilating yeast Candida tropicalis: gene analysis and characterization". J. Biochem. 107 (2): 262–266. doi:10.1093/oxfordjournals.jbchem.a123036. PMID 2361956.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f g h Dunn, M. F.; Ramírez-Trujillo, J. A.; Hernández-Lucas, I. (2009). "Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis". Microbiology. 155 (10): 3166–3175. doi:10.1099/mic.0.030858-0. PMID 19684068.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) CS1 maint: unflagged free DOI (link) - ^ Garrett R and Grisham CN (2008). Biochemistry. Brooks Cole. p. 588. ISBN 9780495109358.
- ^ Cozzone, AJ (1998). "Regulation of acetate metabolism by protein phosphorylation in enteric bacteria". Annu. Rev. Microbiol. 52: 127–164. doi:10.1146/annurev.micro.52.1.127. PMID 9891796.
- ^ Rehman, A.; McFadden, B. A. (1997). "Lysine 194 is functional in isocitrate lyase from Escherichia coli". Curr. Microbiol. 35 (1): 14–17. doi:10.1007/s002849900203. PMID 9175553.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) - ^ a b Eastmond, P. J.; Graham, I. A. (2001). "Re-examining the role of the glyoxylate cycle in oilseeds". Trends Plant Sci. 6 (2): 72–78. doi:10.1016/s1360-1385(00)01835-5. PMID 11173291.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) - ^ a b Kondrashov, F. A.; Koonin, E. V.; Morgunov, I. G.; Finogenova, T. V.; Kondrashova, M. N. (23). "Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation". Biol Direct. 1 (31): 31. doi:10.1186/1745-6150-1-31. PMC 1630690. PMID 17059607.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Kornberg, HL (18). "Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle". Nature. 179 (4568): 988–991. doi:10.1038/179988a0. PMID 134307066.
{{cite journal}}: Check|pmid=value (help); Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Idnurm, A.; Howlett, B. J. (2002). "Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus)". Eukaryot Cell. 1 (5): 719–724. doi:10.1128/EC.1.5.719-724.2002. PMC 126752. PMID 12455691.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) - ^ Lorenz, M. C.; Bender, J. A.; Fink, G. R. (2004). "Fink GR". Eukaryot Cell. 3 (5): 1076–1087. doi:10.1128/EC.3.5.1076-1087.2004. PMC 522606. PMID 15470236.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: date and year (link) - ^ Srivastava, V (May 2008). "Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice". Tuberculosis (Edinb). 88 (3): 171–177. doi:10.1016/j.tube.2007.10.002. PMID 18054522.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: date and year (link) - ^ Muñoz-Elías, EJ (Jun 2005). "Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence". Nat Med. 11 (6): 638–644. doi:10.1038/nm1252. PMC 1464426. PMID 15895072.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: date and year (link)
Further reading
[edit]- McFadden BA and Howes WV (1963). "Crystallisation and some properties of isocitrate lyase from Pseudomonas indigofera". J. Biol. Chem. 238 (5): 1737–1742. doi:10.1016/S0021-9258(18)81129-2.
- Shiio I, Shiio T and McFadden BA (1965). "Isocitrate lyase from Pseudomonas indigofera. I. Preparation, amino acid composition and molecular weight". Biochim. Biophys. Acta. 96: 114–122. doi:10.1016/0005-2787(65)90615-5. PMID 14285253.
- VICKERY HB (1962). "A suggested new nomenclature for the isomers of isocitric acid". J. Biol. Chem. 237 (6): 1739–41. doi:10.1016/S0021-9258(19)73928-3. PMID 13925783.