Microbiome: Difference between revisions
Epipelagic (talk | contribs) m remove unsourced addition |
Epipelagic (talk | contribs) →Background: replace table image with table text Tags: use of predatory open access journal Disambiguation links added |
||
| Line 18: | Line 18: | ||
==Background== |
==Background== |
||
[[File:Microbiome history of method innovations and discoveries.png|thumb|upright=2.6|right| {{center|'''Timeline of microbiome research from seventeenth century''' <ref name=Berg2020 />}} {{align|right|[https://springernature.figshare.com/articles/journal_contribution/Additional_file_1_of_Microbiome_definition_re-visited_old_concepts_and_new_challenges/12592671/1 List of sources used for this figure]}}]] |
|||
Microbiome research originated in microbiology and started back in the seventeenth century. The development of new techniques and equipment has boosted microbiological research and caused paradigm shifts in understanding health and disease. Since infectious diseases have affected human populations throughout most of history, [[medical microbiology]] was the earliest focus of research and public interest. Additionally, [[food microbiology]] is an old field of empirical applications. The development of the first [[microscope]]s allowed the discovery of a new, unknown world and led to the identification of [[microorganism]]s.<ref name=Berg2020 /> |
Microbiome research originated in microbiology and started back in the seventeenth century. The development of new techniques and equipment has boosted microbiological research and caused paradigm shifts in understanding health and disease. Since infectious diseases have affected human populations throughout most of history, [[medical microbiology]] was the earliest focus of research and public interest. Additionally, [[food microbiology]] is an old field of empirical applications. The development of the first [[microscope]]s allowed the discovery of a new, unknown world and led to the identification of [[microorganism]]s.<ref name=Berg2020 /> |
||
| Line 26: | Line 24: | ||
{{clear}} |
{{clear}} |
||
[[File:Microbiome paradigm shifts.png|thumb|upright=1.6| |
[[File:Microbiome paradigm shifts.png|thumb|upright=1.6|right| {{center|'''Paradigm shift'''}} Highlighting the shift of the paradigm from microbes as unsocial organisms causing diseases to the holistic view of microorganisms being the center of the [[One Health Concept]]: positively interconnecting all areas of human lives.<ref name=Berg2020 />]] |
||
However, comprehensive research over the past century has shown only a small proportion of microorganisms are associated with disease or pathogenicity. The overwhelming majority of [[microbe]]s are essential for [[ecosystem|ecosystem functioning]] and known for beneficial interactions with other microbes as well as macroorganisms. At the end of the nineteenth century, [[microbial ecology]] started with the pioneering work by [[Martinus W. Beijerinck]] and [[Sergei Winogradsky]]. The newly established science of [[environmental microbiology]] resulted in another paradigm shift: microorganisms are everywhere in natural environments, often associated with [[Host (biology)|hosts]] and, for the first time, beneficial effects on their hosts were reported.<ref>Hiltner L. (1902) "Die Keimungsverhältnisse der Leguminosensamen und ihre Beeinflussung durch Organismenwirkung". In: Parey P and Springer J (Eds.) ''Arb Biol Abt Land u Forstw K Gsndhtsamt'', '''3''', Berlin. Pages 1-545.</ref><ref>Metchnikoff E. The prolongation of life: optimistic studies. GP Putnam's Sons; 1908.</ref><ref name=Berg2020 /> |
However, comprehensive research over the past century has shown only a small proportion of microorganisms are associated with disease or pathogenicity. The overwhelming majority of [[microbe]]s are essential for [[ecosystem|ecosystem functioning]] and known for beneficial interactions with other microbes as well as macroorganisms. At the end of the nineteenth century, [[microbial ecology]] started with the pioneering work by [[Martinus W. Beijerinck]] and [[Sergei Winogradsky]]. The newly established science of [[environmental microbiology]] resulted in another paradigm shift: microorganisms are everywhere in natural environments, often associated with [[Host (biology)|hosts]] and, for the first time, beneficial effects on their hosts were reported.<ref>Hiltner L. (1902) "Die Keimungsverhältnisse der Leguminosensamen und ihre Beeinflussung durch Organismenwirkung". In: Parey P and Springer J (Eds.) ''Arb Biol Abt Land u Forstw K Gsndhtsamt'', '''3''', Berlin. Pages 1-545.</ref><ref>Metchnikoff E. The prolongation of life: optimistic studies. GP Putnam's Sons; 1908.</ref><ref name=Berg2020 /> |
||
| Line 37: | Line 35: | ||
{{clear}} |
{{clear}} |
||
{| class="wikitable mw-collapsible autocollapse" |
|||
|- |
|||
! colspan=5 width=700px align=center style=background:#ddf8f8 | Timeline of microbiome research from the seventeenth century to the present{{hsp}}<ref name=Berg2020 /> |
|||
|- |
|||
! {{align|right|Technological developments}} |
|||
! width=70px | Year |
|||
! {{align|left|Scientific discoveries}} |
|||
! Scientists |
|||
! Sources |
|||
|- |
|||
| align=right valign=top rowspan=4 | [[microscopy]] |
|||
| align=center valign=top | 1670 |
|||
| valign=top | [[Van Leeuwenhoek's microscopic discovery of microbial life|discovery of microorganisms]] |
|||
| align=center style=background:#edfcfc | [[Antonie van Leeuwenhoek]]<br /><small>father of microbiology</small> |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1098/rstb.2014.0344|title = The unseen world: Reflections on Leeuwenhoek (1677) 'Concerning little animals'|year = 2015|last1 = Lane|first1 = Nick|journal = Philosophical Transactions of the Royal Society B: Biological Sciences|volume = 370|issue = 1666|pmid = 25750239|pmc = 4360124}}</ref> |
|||
|- |
|||
| align=center | 1729 |
|||
| [[:it:Pier Antonio Micheli#Manoscritti ed erbario|classification of plants and fungi]] |
|||
| align=center style=background:#edfcfc | [[Pier Antonio Micheli]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1080/00837792.2016.1147210|title = Pier Antonio Micheli (1679–1737) and Carl Linnaeus (1707–1778)|year = 2016|last1 = Jarvis|first1 = Charles E.|journal = Webbia|volume = 71|pages = 1–24|s2cid = 88308313}}</ref> |
|||
|- |
|||
| align=center | 1796 |
|||
| [[Vaccination#History|first vaccination]] |
|||
| align=center style=background:#edfcfc | [[Edward Jenner]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1080/08998280.2005.11928028|title = Edward Jenner and the History of Smallpox and Vaccination|year = 2005|last1 = Riedel|first1 = Stefan|journal = Baylor University Medical Center Proceedings|volume = 18|issue = 1|pages = 21–25|pmid = 16200144|pmc = 1200696}}</ref> |
|||
|- |
|||
| align=center | 1837 |
|||
| [[Theodor Schwann#Yeast, fermentation, and spontaneous generation|yeast in alcoholic fermentation]] |
|||
| align=center style=background:#edfcfc | [[Charles Cagniard de la Tour|Charles de la Tour]]<br />[[Friedrich Traugott Kützing|Friedrich Kützing]]<br />[[Theodor Schwann]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1080/09571269308717966|title = Origin and domestication of the wine yeast ''Saccharomyces'' cerevisiae|year = 1993|last1 = Martini|first1 = Alessandro|journal = Journal of Wine Research|volume = 4|issue = 3|pages = 165–176}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=6 | [[Microbiological culture|cultivation based approaches]] |
|||
| align=center | 1855<br />-1857 |
|||
| [[Pasteurization#History|pasteurisation]], [[Fermentation#History of the use of fermentation|fermentation]],<br />[[Rabies vaccine#History|vaccine against rabies]] |
|||
| align=center style=background:#edfcfc | [[Louis Pasteur]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal | last=Berche | first=P. | title=Louis Pasteur, from crystals of life to vaccination | journal=Clinical Microbiology and Infection | publisher=Elsevier BV | volume=18 | year=2012 | issn=1198-743X | doi=10.1111/j.1469-0691.2012.03945.x | pages=1–6}}</ref> |
|||
|- |
|||
| align=center | 1875 |
|||
| [[Bacterial taxonomy#Early formal classifications|foundation for bacteriological taxonomy]] |
|||
| align=center style=background:#edfcfc | [[Ferdinand Cohn]] |
|||
| align=center style=background:#edfcfc | |
|||
|- |
|||
| align=center | 1884 |
|||
| [[Koch's postulates]] |
|||
| align=center style=background:#edfcfc | [[Robert Koch]] |
|||
| align=center style=background:#edfcfc | <ref>Evans, A.S. (1976) [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2595276/ "Causation and disease: the Henle-Koch postulates revisited]. ''The Yale journal of biology and medicine'', '''49'''(2): 175.</ref> |
|||
|- |
|||
| align=center | 1888 |
|||
| [[Microbial ecology#History|start of microbial ecology]]<br /><small>[[nitrification]], [[nitrogen-fixation]], [[soil microbiology]], life cycle</small> |
|||
| align=center style=background:#edfcfc | [[Sergei Winogradsky]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1111/j.1574-6976.2011.00299.x|title = Sergei Winogradsky: A founder of modern microbiology and the first microbial ecologist|year = 2012|last1 = Dworkin|first1 = Martin|last2 = Gutnick|first2 = David|journal = FEMS Microbiology Reviews|volume = 36|issue = 2|pages = 364–379|pmid = 22092289}}</ref> |
|||
|- |
|||
| align=center | 1892 |
|||
| [[Tobacco mosaic virus#History|tobacco mosaic virus extraction from leaves]] |
|||
| align=center style=background:#edfcfc | [[Dmitri Ivanovsky]]<br />[[Martinus Beijerinck]] |
|||
| align=center style=background:#edfcfc | |
|||
|- |
|||
| align=center | 1904 |
|||
| [[Rhizosphere|concept of the rhizosphere]] |
|||
| align=center style=background:#edfcfc | [[:de:Lorenz Hiltner|Lorenz Hiltner]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1007/s11104-007-9514-z|title = Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research|year = 2008|last1 = Hartmann|first1 = Anton|last2 = Rothballer|first2 = Michael|last3 = Schmid|first3 = Michael|journal = Plant and Soil|volume = 312|issue = 1–2|pages = 7–14|s2cid = 4419735}}</ref><ref>{{cite journal |doi = 10.1007/s11104-007-9514-z|title = Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research|year = 2008|last1 = Hartmann|first1 = Anton|last2 = Rothballer|first2 = Michael|last3 = Schmid|first3 = Michael|journal = Plant and Soil|volume = 312|issue = 1–2|pages = 7–14|s2cid = 4419735}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=1 | [[fluorescence microscopy]] |
|||
| align=center | 1911 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite web |url=http://nobelprize.org/educational_games/physics/microscopes/fluorescence/ |title=The Fluorescence Microscope |publisher=[[The Nobel Foundation]] |work=Microscopes—Help Scientists Explore Hidden Worlds |access-date=2008-09-28}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=4 | [[History of mass spectrometry|mass spectrometry]] |
|||
| align=center | 1919 |
|||
| |
|||
| align=center style=background:#edfcfc| [[Francis Aston]] |
|||
| align=center style=background:#edfcfc | <ref>Borman, S., Russell, H. and Siuzdak, G., (2003) "A Mass Spec Timeline Developing techniques to measure mass has been a Nobel pursuit. ''Todays Chemist at Work'', '''12'''(9): 47–50.</ref> |
|||
|- |
|||
| align=center | 1922 |
|||
| [[chemolithotrophy]] |
|||
| align=center style=background:#edfcfc | [[Sergei Winogradsky]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1126/science.118.3054.36|title = Sergei Nikolaevitch Winogradsky: 1856-1953|year = 1953|last1 = Waksman|first1 = Selman A.|journal = Science|volume = 118|issue = 3054|pages = 36–37|pmid = 13076173|bibcode = 1953Sci...118...36W}}</ref> |
|||
|- |
|||
| align=center | 1928 |
|||
| [[Griffith's experiment|transformation of genetic information<br />to offspring]] |
|||
| align=center style=background:#edfcfc | [[Frederick Griffith]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1017/S0022172400031879|title = The Significance of Pneumococcal Types|year = 1928|last1 = Griffith|first1 = Fred|journal = Journal of Hygiene|volume = 27|issue = 2|pages = 113–159|pmid = 20474956|pmc = 2167760}}</ref><ref>Hayes, W. (1966) "Genetic Transformation: a Retrospective Appreciation", First Griffith Memorial Lecture. ''Microbiology'', '''45'''(3): 385-397.</ref> |
|||
|- |
|||
| align=center | 1928 |
|||
| [[History of penicillin|discovery of antibiotics]] |
|||
| align=center style=background:#edfcfc | [[Alexander Fleming]] |
|||
| align=center style=background:#edfcfc | <ref>American Chemical Society (1999) [https://www.acs.org/content/dam/acsorg/education/whatischemistry/landmarks/flemingpenicillin/the-discovery-and-development-of-penicillin-commemorative-booklet.pdf ''Discovery and Development of Penicillin, 1928–1945'']. International Historic Chemical Landmarks, The Alexander Fleming Laboratory Museum, London.</ref> |
|||
|- |
|||
| align=right valign=top rowspan=4 | [[Scanning electron microscope|scanning electron microscopy]] |
|||
| align=center | 1931<br />-1938 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1002/anie.198705953|title = The Development of the Electron Microscope and of Electron Microscopy(Nobel Lecture)|year = 1987|last1 = Ruska|first1 = Ernst|journal = Angewandte Chemie International Edition in English|volume = 26|issue = 7|pages = 595–605}}</ref> |
|||
|- |
|||
| align=center | 1944 |
|||
| [[History of DNA biochemistry|DNA as carrier of genetic information]] |
|||
| align=center style=background:#edfcfc | [[Oswald Avery]]<br />[[Colin Macleod]]<br />[[Maclyn McCarty]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1084/jem.149.2.297|title = Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III|year = 1979|last1 = Avery|first1 = O. T.|last2 = MacLeod|first2 = C. M.|last3 = McCarty|first3 = M.|journal = Journal of Experimental Medicine|volume = 149|issue = 2|pages = 297–326|pmid = 33226|pmc = 2184805}}</ref> |
|||
|- |
|||
| align=center | 1946 |
|||
| [[Transduction (genetics)|"sexual reproduction" of bacteria]] |
|||
| align=center style=background:#edfcfc | [[Joshua Lederberg]]<br />[[Edward Tatum]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1007/s10739-017-9493-8|title = The Experimental Study of Bacterial Evolution and Its Implications for the Modern Synthesis of Evolutionary Biology|year = 2018|last1 = o'Malley|first1 = Maureen A.|journal = Journal of the History of Biology|volume = 51|issue = 2|pages = 319–354|pmid = 28980196|s2cid = 4055566}}</ref> |
|||
|- |
|||
| align=center | 1953 |
|||
| [[Nucleic acid double helix|3D-double-helix structure]]{{hsp}}<ref>{{cite journal |doi = 10.1038/nsb0403-247|title = The double helix: A tale of two puckers|year = 2003|last1 = Rich|first1 = Alexander|journal = Nature Structural & Molecular Biology|volume = 10|issue = 4|pages = 247–249|pmid = 12660721|s2cid = 6089989}}</ref> |
|||
| align=center style=background:#edfcfc | [[James Watson]]<br />[[Francis Crick]] |
|||
| align=center style=background:#edfcfc | |
|||
|- |
|||
| align=right valign=top rowspan=1 | ''in situ'' hybridisation iSIS |
|||
| align=center | 1969 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref name=Cassidy2014>{{cite journal |doi = 10.1016/j.ymeth.2014.04.006|title = Developments in in situ hybridisation|year = 2014|last1 = Cassidy|first1 = Andrew|last2 = Jones|first2 = Julia|journal = Methods|volume = 70|issue = 1|pages = 39–45|pmid = 24747923}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=1 | [[HPLC]] |
|||
| align=center | 1970s |
|||
| [[central dogma of molecular biology]]{{hsp}}<ref>{{cite journal |doi = 10.1038/227561a0|title = Central Dogma of Molecular Biology|year = 1970|last1 = Crick|first1 = Francis|journal = Nature|volume = 227|issue = 5258|pages = 561–563|pmid = 4913914|bibcode = 1970Natur.227..561C|s2cid = 4164029}}</ref> |
|||
| align=center style=background:#edfcfc | [[Francis Crick]] |
|||
| align=center style=background:#edfcfc |<ref>{{cite book | last=Meyer | first=Veronika | title=Practical high-performance liquid chromatography | url=https://books.google.com/books?id=ODRYwLsJy3AC | publisher=Wiley | publication-place=Hoboken, N.J | year=2013 | isbn=978-1-118-68134-3 | oclc=864917338}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=1 | [[Array hybridization|DNA array]]/[[Colony hybridization|colony hybridisation]] |
|||
| align=center | 1975 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1073/pnas.72.10.3961|title = Colony hybridization: A method for the isolation of cloned DNAs that contain a specific gene|year = 1975|last1 = Grunstein|first1 = M.|last2 = Hogness|first2 = D. S.|journal = Proceedings of the National Academy of Sciences|volume = 72|issue = 10|pages = 3961–3965|pmid = 1105573|pmc = 433117|bibcode = 1975PNAS...72.3961G|doi-access = free}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=4 | [[Sanger sequencing]] |
|||
| align=center | 1977 |
|||
| |
|||
| align=center style=background:#edfcfc | [[Frederick Sanger]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1073/pnas.74.12.5463|title = DNA sequencing with chain-terminating inhibitors|year = 1977|last1 = Sanger|first1 = F.|last2 = Nicklen|first2 = S.|last3 = Coulson|first3 = A. R.|journal = Proceedings of the National Academy of Sciences|volume = 74|issue = 12|pages = 5463–5467|pmid = 271968|pmc = 431765|bibcode = 1977PNAS...74.5463S|doi-access = free}}</ref><ref>{{cite journal |doi = 10.1016/j.ygeno.2015.11.003|title = The sequence of sequencers: The history of sequencing DNA|year = 2016|last1 = Heather|first1 = James M.|last2 = Chain|first2 = Benjamin|journal = Genomics|volume = 107|pages = 1–8|pmid = 26554401|s2cid = 27846422}}</ref> |
|||
|- |
|||
| align=center | 1977 |
|||
| discovery of [[Archaea]] |
|||
| align=center style=background:#edfcfc | [[Carl Woese]]<br />[[George E. Fox]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1073/pnas.74.11.5088|title = Phylogenetic structure of the prokaryotic domain: The primary kingdoms|year = 1977|last1 = Woese|first1 = C. R.|last2 = Fox|first2 = G. E.|journal = Proceedings of the National Academy of Sciences|volume = 74|issue = 11|pages = 5088–5090|pmid = 270744|pmc = 432104|bibcode = 1977PNAS...74.5088W|doi-access = free}}</ref><ref>{{cite journal |doi = 10.1038/nrmicro.2017.133|title = Archaea and the origin of eukaryotes|year = 2017|last1 = Eme|first1 = Laura|last2 = Spang|first2 = Anja|last3 = Lombard|first3 = Jonathan|last4 = Stairs|first4 = Courtney W.|last5 = Ettema|first5 = Thijs J. G.|journal = Nature Reviews Microbiology|volume = 15|issue = 12|pages = 711–723|pmid = 29123225|s2cid = 8666687}}</ref> |
|||
|- |
|||
| align=center | 1977 |
|||
| first [[full genome sequence]] of a [[Virus#Genome|virus]] |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1038/260500a0|title = Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene|year = 1976|last1 = Fiers|first1 = W.|last2 = Contreras|first2 = R.|last3 = Duerinck|first3 = F.|last4 = Haegeman|first4 = G.|last5 = Iserentant|first5 = D.|last6 = Merregaert|first6 = J.|last7 = Min Jou|first7 = W.|last8 = Molemans|first8 = F.|last9 = Raeymaekers|first9 = A.|last10 = Van Den Berghe|first10 = A.|last11 = Volckaert|first11 = G.|last12 = Ysebaert|first12 = M.|journal = Nature|volume = 260|issue = 5551|pages = 500–507|pmid = 1264203|bibcode = 1976Natur.260..500F|s2cid = 4289674}}</ref> |
|||
|- |
|||
| align=center | 1982 |
|||
| [[Prion#History|discovery of prions]] |
|||
| align=center style=background:#edfcfc | [[Stanley B. Prusiner]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1126/science.6801762 |
|||
|title = Novel Proteinaceous Infectious Particles Cause Scrapie |
|||
|year = 1982 |
|||
|last1 = Prusiner |
|||
|first1 = Stanley B. |
|||
|journal = Science |
|||
|volume = 216 |
|||
|issue = 4542 |
|||
|pages = 136–144 |
|||
|pmid = 6801762 |
|||
|bibcode = 1982Sci...216..136P |
|||
}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=1 | [[PCR|PCR technique]] |
|||
| align=center | 1983 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>Mullis, K.B. (1990) [https://www.jstor.org/stable/24996713 "The unusual origin of the polymerase chain reaction"]. ''Scientific American'', '''262'''(4): 56–65.</ref><ref>{{cite journal |doi = 10.1038/nbt0993-1026|title = Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions|year = 1993|last1 = Higuchi|first1 = Russell|last2 = Fockler|first2 = Carita|last3 = Dollinger|first3 = Gavin|last4 = Watson|first4 = Robert|journal = Nature Biotechnology|volume = 11|issue = 9|pages = 1026–1030|pmid = 7764001|s2cid = 5714001}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=2 | [[fluorescence in situ hybridisation|fluorescence ''in situ'' hybridisation]] |
|||
| align=center | 1988 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc |<ref name=Cassidy2014 /><ref>{{cite journal |doi = 10.1016/0092-8674(88)90530-2|title = Sensitive, high-resolution chromatin and chromosome mapping in situ: Presence and orientation of two closely integrated copies of EBV in a lymphoma line|year = 1988|last1 = Bentleylawrence|first1 = J.|last2 = Villnave|first2 = C. A.|last3 = Singer|first3 = R. H.|journal = Cell|volume = 52|issue = 1|pages = 51–61|pmid = 2830981|s2cid = 17565963}}</ref> |
|||
|- |
|||
| align=center | 1991 |
|||
| [[Hologenome theory of evolution|theory of the holobiont]] |
|||
| align=center style=background:#edfcfc | [[Eugene Rosenberg]]<br />Ilana Zilber-Rosenberg |
|||
| align=center style=background:#edfcfc | <ref>{{cite book | last=Margulis | first=Lynn | title=Symbiosis as a source of evolutionary innovation : speciation and morphogenesis | url= https://books.google.com/books?id=3sKzeiHUIUQC | publisher=MIT Press | publication-place=Cambridge, Mass | year=1991 | isbn=978-0-262-13269-5 | oclc=22597587}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=2 | quantitative real-time [[PCR]] |
|||
| align=center | 1993 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | |
|||
|- |
|||
| align=center | 1993 |
|||
| the complex structure of [[biofilm]]s |
|||
| align=center style=background:#edfcfc | [[:de:Hans-Curt Flemming|Hans-Curt Flemming]] |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.2166/wst.1993.0528|title = Biofilms and Environmental Protection|year = 1993|last1 = Flemming|first1 = Hans-Curt|journal = Water Science and Technology|volume = 27|issue = 7–8|pages = 1–10}}</ref><ref>{{cite book | author=Flemming | title=Biofilm highlights | url=https://books.google.com/books?id=1azigYOg8woC | publisher=Springer-Verlag Berlin Heidelberg | publication-place=Heidelberg New York | year=2011 | isbn=978-3-642-19940-0 | oclc=769756150}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=2 | full cycle [[Ribosomal RNA|rRNA approach]] |
|||
| align=center | 1995 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1128/mr.59.1.143-169.1995|title = Phylogenetic identification and in situ detection of individual microbial cells without cultivation|year = 1995|last1 = Amann|first1 = R. I.|last2 = Ludwig|first2 = W.|last3 = Schleifer|first3 = K. H.|journal = Microbiological Reviews|volume = 59|issue = 1|pages = 143–169|pmid = 7535888|pmc = 239358}}</ref> |
|||
|- |
|||
| align=center | 1995 |
|||
| first full [[genome]] sequencing<br />of ''[[Haemophilus influenzae]]'' |
|||
| align=center style=background:#edfcfc | [[Craig Venter]]<br />and colleagues |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1126/science.7542800 |
|||
|title = Whole-Genome Random Sequencing and Assembly of Haemophilus influenzae Rd |
|||
|year = 1995 |
|||
|last1 = Fleischmann |
|||
|first1 = Robert D. |
|||
|last2 = Adams |
|||
|first2 = Mark D. |
|||
|last3 = White |
|||
|first3 = Owen |
|||
|last4 = Clayton |
|||
|first4 = Rebecca A. |
|||
|last5 = Kirkness |
|||
|first5 = Ewen F. |
|||
|last6 = Kerlavage |
|||
|first6 = Anthony R. |
|||
|last7 = Bult |
|||
|first7 = Carol J. |
|||
|last8 = Tomb |
|||
|first8 = Jean-Francois |
|||
|last9 = Dougherty |
|||
|first9 = Brian A. |
|||
|last10 = Merrick |
|||
|first10 = Joseph M. |
|||
|last11 = McKenney |
|||
|first11 = Keith |
|||
|last12 = Sutton |
|||
|first12 = Granger |
|||
|last13 = Fitzhugh |
|||
|first13 = Will |
|||
|last14 = Fields |
|||
|first14 = Chris |
|||
|last15 = Gocayne |
|||
|first15 = Jeannine D. |
|||
|last16 = Scott |
|||
|first16 = John |
|||
|last17 = Shirley |
|||
|first17 = Robert |
|||
|last18 = Liu |
|||
|first18 = Li-lng |
|||
|last19 = Glodek |
|||
|first19 = Anna |
|||
|last20 = Kelley |
|||
|first20 = Jenny M. |
|||
|last21 = Weidman |
|||
|first21 = Janice F. |
|||
|last22 = Phillips |
|||
|first22 = Cheryl A. |
|||
|last23 = Spriggs |
|||
|first23 = Tracy |
|||
|last24 = Hedblom |
|||
|first24 = Eva |
|||
|last25 = Cotton |
|||
|first25 = Matthew D. |
|||
|last26 = Utterback |
|||
|first26 = Teresa R. |
|||
|last27 = Hanna |
|||
|first27 = Michael C. |
|||
|last28 = Nguyen |
|||
|first28 = David T. |
|||
|last29 = Saudek |
|||
|first29 = Deborah M. |
|||
|last30 = Brandon |
|||
|first30 = Rhonda C. |
|||
|journal = Science |
|||
|volume = 269 |
|||
|issue = 5223 |
|||
|pages = 496–512 |
|||
|pmid = 7542800 |
|||
|bibcode = 1995Sci...269..496F |
|||
|display-authors = 1 |
|||
}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=2 | [[next-generation sequencing]] |
|||
| align=center | 2005 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite book |doi = 10.5772/61964|chapter = Next-Generation Sequencing — an Overview of the History, Tools, and "Omic" Applications|title = Next Generation Sequencing - Advances, Applications and Challenges|year = 2016|last1 = Kulski|first1 = Jerzy K.|isbn = 978-953-51-2240-1}}</ref> |
|||
|- |
|||
| align=center | 2005 |
|||
| HMP: [[Human Microbiome Project]] |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1101/gr.138297.112|title = CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome|year = 2012|last1 = Stern|first1 = A.|last2 = Mick|first2 = E.|last3 = Tirosh|first3 = I.|last4 = Sagy|first4 = O.|last5 = Sorek|first5 = R.|journal = Genome Research|volume = 22|issue = 10|pages = 1985–1994|pmid = 22732228|pmc = 3460193}}</ref> |
|||
|- |
|||
| align=right valign=top rowspan=3 | [[third-generation sequencing]] |
|||
| align=center | 2008 |
|||
| |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1093/hmg/ddq416|title = A window into third-generation sequencing|year = 2010|last1 = Schadt|first1 = E. E.|last2 = Turner|first2 = S.|last3 = Kasarskis|first3 = A.|journal = Human Molecular Genetics|volume = 19|issue = R2|pages = R227–R240|pmid = 20858600}}</ref> |
|||
|- |
|||
| align=center | 2008 |
|||
| TerraGenome:<br />Reference Soil Metagenome Project |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.1038/nrmicro2119|title = Terra ''Genome'': A consortium for the sequencing of a soil metagenome|year = 2009|last1 = Vogel|first1 = Timothy M.|last2 = Simonet|first2 = Pascal|last3 = Jansson|first3 = Janet K.|last4 = Hirsch|first4 = Penny R.|last5 = Tiedje|first5 = James M.|last6 = Van Elsas|first6 = Jan Dirk|last7 = Bailey|first7 = Mark J.|last8 = Nalin|first8 = Renaud|last9 = Philippot|first9 = Laurent|journal = Nature Reviews Microbiology|volume = 7|issue = 4|page = 252|s2cid = 2144462}}</ref> |
|||
|- |
|||
| align=center | 2010 |
|||
| [[Earth Microbiome Project]] |
|||
| style=background:#edfcfc | |
|||
| align=center style=background:#edfcfc | <ref>{{cite journal |doi = 10.4056/aigs.1443528|title = The Earth Microbiome Project: Meeting report of the "1st EMP meeting on sample selection and acquisition" at Argonne National Laboratory October 6th 2010|year = 2010|last1 = Gilbert|first1 = Jack A.|last2 = Meyer|first2 = Folker|last3 = Jansson|first3 = Janet|last4 = Gordon|first4 = Jeff|last5 = Pace|first5 = Norman|last6 = Tiedje|first6 = James|last7 = Ley|first7 = Ruth|last8 = Fierer|first8 = Noah|last9 = Field|first9 = Dawn|last10 = Kyrpides|first10 = Nikos|last11 = Glöckner|first11 = Frank-Oliver|last12 = Klenk|first12 = Hans-Peter|last13 = Wommack|first13 = K. Eric|last14 = Glass|first14 = Elizabeth|last15 = Docherty|first15 = Kathryn|last16 = Gallery|first16 = Rachel|last17 = Stevens|first17 = Rick|last18 = Knight|first18 = Rob|journal = Standards in Genomic Sciences|volume = 3|issue = 3|pages = 249–253|pmid = 21304728|pmc = 3035312}}</ref> |
|||
|- |
|||
|} |
|||
==Defining the microbiome== |
==Defining the microbiome== |
||
Revision as of 07:41, 14 November 2021

| Part of a series on |
| Microbiomes |
|---|
 |
The word microbiome (from the Greek micro meaning "small" and bíos meaning "life") was first used by J.L. Mohr in 1952 in The Scientific Monthly to mean the microorganisms found in a specific environment.[2][3] It was defined in 1988 by Whipps et al. as "a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theatre of activity".[4]
In 2020, an international panel of experts published the outcome of their discussions on the definition of the microbiome.[1] They proposed a definition of the microbiome based on a revival of the "compact, clear, and comprehensive description of the term" as originally provided by Whipps et al., but supplemented with two explanatory sentences.[1]
The first explanatory sentence pronounces the dynamic character of the microbiome:
- The microbiome is defined as a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The microbiome not only refers to the microorganisms involved but also encompass their theatre of activity, which results in the formation of specific ecological niches. The microbiome, which forms a dynamic and interactive micro-ecosystem prone to change in time and scale, is integrated in macro-ecosystems including eukaryotic hosts, and here crucial for their functioning and health.[1]
The second explanatory sentence clearly separates the term microbiota from the term microbiome:
- The microbiota consists of the assembly of microorganisms belonging to different kingdoms (Prokaryotes [Bacteria, Archaea], Eukaryotes [e.g., Protozoa, Fungi, and Algae]), while their theatre of activity includes microbial structures, metabolites, mobile genetic elements (such as transposons, phages, and viruses), and relic DNA embedded in the environmental conditions of the habitat.[1]
Secondary metabolites play an essential role in mediating complex interspecies interactions and ensure survival in competitive environments. Quorum sensing induced by small molecules allows bacteria to control cooperative activities and adapts their phenotypes to the biotic environment, resulting, e.g., in cell-cell adhesion or biofilm formation. Direct interspecies electron transfer (DIET) is an important mechanism for communication in most anaerobic ecosystems. In addition, volatile compounds can act as long-term messengers for cross-kingdom communication over long distances.
Background
Microbiome research originated in microbiology and started back in the seventeenth century. The development of new techniques and equipment has boosted microbiological research and caused paradigm shifts in understanding health and disease. Since infectious diseases have affected human populations throughout most of history, medical microbiology was the earliest focus of research and public interest. Additionally, food microbiology is an old field of empirical applications. The development of the first microscopes allowed the discovery of a new, unknown world and led to the identification of microorganisms.[1]
Access to the previously invisible world opened the eyes and the minds of the researchers of the seventeenth century. Antonie van Leeuwenhoek investigated diverse bacteria of various shapes, fungi, and protozoa, which he called animalcules, mainly from water, mud, and dental plaque samples, and discovered biofilms as a first indication of microorganisms interacting within complex communities. Robert Koch's explanation of the origin of human and animal diseases as a consequence of microbial infection and development of the concept of pathogenicity was an important milestone in microbiology. These findings shifted the focus of the research community and the public on the role of microorganisms as disease-forming agents that needed to be eliminated.[1]

However, comprehensive research over the past century has shown only a small proportion of microorganisms are associated with disease or pathogenicity. The overwhelming majority of microbes are essential for ecosystem functioning and known for beneficial interactions with other microbes as well as macroorganisms. At the end of the nineteenth century, microbial ecology started with the pioneering work by Martinus W. Beijerinck and Sergei Winogradsky. The newly established science of environmental microbiology resulted in another paradigm shift: microorganisms are everywhere in natural environments, often associated with hosts and, for the first time, beneficial effects on their hosts were reported.[5][6][1]
Subsequently, the concept that microorganisms exist as single cells began to change as it became increasingly obvious that microbes occur within complex assemblages in which species interactions and communication are critical to population dynamics and functional activities.[7] Discovery of DNA, the development of sequencing technologies, PCR, and cloning techniques enabled the investigation of microbial communities using cultivation-independent, DNA and RNA-based approaches.[8][1]
A further important step was the introduction of phylogenetic markers such as the 16S rRNA gene for microbial community analysis by Carl Woese and George E. Fox in 1977.[9] Today, we are able to barcode bacteria, archaea, fungi, algae, and protists in their natural habitats, e.g., by targeting their 16S and 18S rRNA genes, internal transcribed spacer (ITS), or, alternatively, specific functional regions of genes coding for specific enzymes.[10][11][12][1]
Another major paradigm shift was initiated at the beginning of this century and continues through today, as new sequencing technologies and accumulated sequence data have highlighted both the ubiquity of microbial communities in association within higher organisms and the critical roles of microbes in human, animal, and plant health.[13] These new possibilities have revolutionized microbial ecology, because the analysis of genomes and metagenomes in a high-throughput manner provides efficient methods for addressing the functional potential of individual microorganisms as well as of whole communities in their natural habitats.[14][15] Multiomics technologies including metatranscriptome, metaproteome and metabolome approaches now provide detailed information on microbial activities in the environment. Based on the rich foundation of data, the cultivation of microbes, which was often ignored or underestimated over the last thirty years, has gained new importance, and high throughput culturomics is now an important part of the toolbox to study microbiomes. The high potential and power of combining multiple "omics" techniques to analyze host-microbe interactions are highlighted in several reviews.[16][17][1]
Defining the microbiome
Microbial communities have commonly been defined as the collection of microorganisms living together. More specifically, microbial communities are defined as multi-species assemblages, in which (micro) organisms interact with each other in a contiguous environment.[60] In 1988, Whipps and colleagues working on the ecology of rhizosphere microorganisms provided the first definition of the term microbiome.[4] They described the microbiome as a combination of the words micro and biome, naming a "characteristic microbial community" in a "reasonably well-defined habitat which has distinct physio-chemical properties" as their "theatre of activity". This definition represents a substantial advancement of the definition of a microbial community, as it defines a microbial community with distinct properties and functions and its interactions with its environment, resulting in the formation of specific ecological niches.[1]
However, many other microbiome definitions have been published in the last few decades. The currently most cited definition by Lederberg[61] describes microbiomes within an ecological context, as a community of commensal, symbiotic, and pathogenic microorganisms within a body space or other environment. Marchesi and Ravel focused in their definition on the genomes and microbial (and viral) gene expression patterns and proteomes in a given environment and its prevailing biotic and abiotic conditions.[62] All these definitions imply that general concepts of macro-ecology could be easily applied to microbe-microbe as well as to microbe-host interactions. However, the extent to which these concepts, developed for macro-eukaryotes, can be applied to prokaryotes with their different lifestyles regarding dormancy, variation of phenotype, and horizontal gene transfer[63] as well as to micro-eukaryotes that is not quite clear. This raises the challenge of considering an entirely novel body of conceptual ecology models and theory for microbiome ecology, particularly in relation to the diverse hierarchies of interactions of microbes with one another and with the host biotic and abiotic environments. Many current definitions fail to capture this complexity and describe the term microbiome as encompassing the genomes of microorganisms only (see table ↓).[1]
| Microbiome definitions[1] | |
|---|---|
| Definition type | Examples |
| Ecological | Definitions based on ecology describe the microbiome following the concepts derived from the ecology of multicellular organisms. The main issue here is that the theories from the macro-ecology do not always fit the rules in the microbial world. |
| |
| Organisms/host-dependent | The host-dependent definitions are based on the microbial interactions with the host. The main gaps here concern the question whether the microbial-host interaction data gained from one host can be transferred to another. The understanding of coevolution and selection in the host-dependent definitions is also underrepresented. |
| |
| Genomic/ method-driven | There is a variety of microbiome definitions available that are driven by the methods applied. Mostly, these definitions rely on DNA sequence-based analysis and describe microbiome as a collective genome of microorganisms in a specific environment. The main bottleneck here is that every new available technology will result in a need for a new definition. |
| |
| Combined | There are some microbiome definitions available that fit several categories with their advantages and disadvantages. |
| |
In 2020, a panel of international experts, organised by the EU-funded MicrobiomeSupport project,[74] published the results of their deliberations on the definition of the microbiome.[1] The panel was composed of about 40 leaders from diverse microbiome areas, and about one hundred further experts from around the world contributed through an online survey. They proposed a definition of the microbiome based on a revival of the compact, clear, and comprehensive description of the term as originally provided by Whipps et al. in 1988,[4] amended with a set of recommendations considering subsequent technological developments and research findings. They clearly separate the terms microbiome and microbiota and provide a comprehensive discussion considering the composition of microbiota, the heterogeneity and dynamics of microbiomes in time and space, the stability and resilience of microbial networks, the definition of core microbiomes, and functionally relevant keystone species as well as co-evolutionary principles of microbe-host and inter-species interactions within the microbiome.[1]
The panel extended the Whipps et al. definition, which contains all important points that are valid even 30 years after its publication in 1988, by two explanatory sentences differentiating the terms microbiome and microbiota and pronouncing its dynamic character, as follows:
- The microbiome is defined as a characteristic microbial community occupying a reasonable well-defined habitat which has distinct physio-chemical properties. The microbiome not only refers to the microorganisms involved but also encompass their theatre of activity, which results in the formation of specific ecological niches. The microbiome, which forms a dynamic and interactive micro-ecosystem prone to change in time and scale, is integrated in macro-ecosystems including eukaryotic hosts, and here crucial for their functioning and health.[1]
- The microbiota consists of the assembly of microorganisms belonging to different kingdoms (prokaryotes (bacteria, archaea), eukaryotes (algae, protozoa, fungi etc), while "their theatre of activity" includes microbial structures, metabolites, mobile genetic elements (such as transposons, phages, and viruses), and relic DNA embedded in the environmental conditions of the habitat.[1]
Microbiota – members of the microbiome
The microbiota comprises all living members forming the microbiome. Most microbiome researchers agree bacteria, archaea, fungi, algae, and small protists should be considered as members of the microbiome.[62][1] The integration of phages, viruses, plasmids, and mobile genetic elements is a more controversial issue in the definition of the microbiome. There is also no clear consensus as to whether extracellular DNA derived from dead cells, so-called "relic DNA", belongs to the microbiome.[75][1] Relic DNA can be up to 40% of the sequenced DNA in soil,[76] and was up to 33% of the total bacterial DNA on average in a broader analysis of habitats with the highest proportion of 80% in some samples.[77] Despite its omnipresence and abundance, relic DNA had a minimal effect on estimates of taxonomic and phylogenetic diversity.[77][1]
When it comes to the use of specific terms, a clear differentiation between microbiome and microbiota helps to avoid the controversy concerning the members of a microbiome.[1] Microbiota is usually defined as the assemblage of living microorganisms present in a defined environment.[62] As phages, viruses, plasmids, prions, viroids, and free DNA are usually not considered as living microorganisms,[78] they do not belong to the microbiota.[1]
The term microbiome, as it was originally postulated by Whipps and coworkers,[4] includes not only the community of the microorganisms but also their "theatre of activity". The latter involves the whole spectrum of molecules produced by the microorganisms, including their structural elements (nucleic acids, proteins, lipids, polysaccharides), metabolites (signalling molecules, toxins, organic, and inorganic molecules), and molecules produced by coexisting hosts and structured by the surrounding environmental conditions. Therefore, all mobile genetic elements, such as phages, viruses, and "relic" and extracellular DNA, should be included in the term microbiome, but are not a part of microbiota. The term microbiome is also sometimes confused with the metagenome. Metagenome is, however, clearly defined as a collection of genomes and genes from the members of a microbiota.[62][1]
Microbiome studies sometimes focus on the behaviour of a specific group of microbiota, generally in relation to or justified by a clear hypothesis. More and more terms like bacteriome, archaeome, mycobiome, or virome have started appearing in the scientific literature, but these terms do not refer to biomes (a regional ecosystem with a distinct assemblage of (micro) organisms, and physical environment often reflecting a certain climate and soil) as the microbiome itself.[1] Consequently, it would be better to use the original terms (bacterial, archaeal, or fungal community). In contrast to the microbiota, which can be studied separately, the microbiome is always composed by all members, which interact with each other, live in the same habitat, and form their ecological niche together. The well-established term virome is derived from virus and genome and is used to describe viral shotgun metagenomes consisting of a collection of nucleic acids associated with a particular ecosystem or holobiont.[79] Viral metagenomes can be suggested as a semantically and scientifically better term.[1]
Microbial networks and interactions

through microbial co-occurrence networks
b) Microbial co-occurrence and co-exclusion networks help visualizing microbial interactions. In such networks, nodes usually represent taxa of microorganisms, and edges represent statistically significant associations between nodes. Green edges usually stay for positive interactions, while red edges visualize negative interactions between the microorganisms.
c) Testing of the hypotheses resulted from the network analyses in relevant model systems is required for a comprehensive study of microbial interactions.[1]
Microbes interact with one another, and these symbiotic interactions have diverse consequences for microbial fitness, population dynamics, and functional capacities within the microbiome.[80] These interactions can either be between microorganisms of the same species or between different species, genera, families, and domains of life. The interactive patterns within these webs may be positive (mutualism, synergism, or commensalism), negative (amensalism [including predation, parasitism, antagonism, or competition]), or neutral—where there is no (or no observed) effect on the functional capacities or fitness of interacting species (see diagram at right) Microbial life strategy concepts (i.e., copiotrophic and oligotrophic strategists and competitor–stress tolerator–ruderals framework) can influence outcomes of interactions.[81] For example, microorganisms competing for the same source can also benefit from each other when competing for the same compound at different trophic levels. Stability of a complex microbial ecosystem depends on trophic interactions for the same substrate at different concentration levels. As of 2020 microbial social adaptations in nature have been understudied.[1] Here molecular markers can provide insight into social adaptations by supporting the theories, e.g., of altruists and cheaters in native microbiomes.[82][1]
Secondary metabolites play an essential role in mediating complex interspecies interactions and ensure survival in competitive environments. Quorum sensing induced by small molecules like n-acyl-homoserine lactones or peptides allows bacteria to control cooperative activities and adapts their phenotypes to the biotic environment, resulting, e.g., in cell-cell adhesion or biofilm formation.[7][83] Direct interspecies electron transfer (DIET) is an important mechanism for communication in most anaerobic ecosystems.[84] In addition, volatile compounds can act as long-term messengers for cross-kingdom communication over long distances.[85] Moreover, the so-called “fungal highways” serve as transportation systems for bacteria [86] as well as for water and nutrients [87] and can therefore play an important role in structuring microbial networks. Despite these examples, communication and interaction within the microbiome remain understudied and would profit from more knowledge on the metabolic interplay of all microbiome members. Here, reductionist experimental models and model microbiomes can help to identify microbes and molecular mechanisms involved in complex interactions.[88][1]
As cells, viruses and macromolecules represent the actors and stage set, so metabolites are the agents that orchestrate the choreography of the theatre. They put on the play, provide the lyrics and let the drama take its course. Metabolites are produced as communication signals, trigger factors, cargo transporters, lubricants, toxins and weapons. To understand the story of the play we need to know the actors and their environment, but most importantly we need to know the metabolites and the interactions that they mediate. This is a challenging task, and the ecological functions of many metabolites still remain obscure. Eavesdropping on microbial crosstalk and elucidating the intricate chemical interactions of microbes goes hand in hand with detailed knowledge about the structures of the metabolites involved and the pathways of their production. – Thomas Böttcher [89]
Assessing microbial functioning

Currently available methods for studying microbiomes, so-called multi-omics, range from high throughput isolation (culturomics) and visualization (microscopy), to targeting the taxonomic composition (metabarcoding), or addressing the metabolic potential (metabarcoding of functional genes, metagenomics) to analyze microbial activity (metatranscriptomics, metaproteomics, metabolomics), as shown in the diagram on the right. Based on metagenome data, microbial genomes can be reconstructed. While first metagenome-assembled genomes were reconstructed from environmental samples,[90] in recent years, several thousands of bacterial genomes were binned without culturing the organisms behind. For example, 154,723 microbial genomes of the global human microbiome were recently reconstructed from 9,428 metagenomes.[91][1]
Computational modeling of microbiomes has been used to compliment experimental methods for investigating microbial function by utilizing multi-omic data to predict complex inter-species and host-species dynamics.[92][93] A popular in silico method is to combine metabolic network models of microbial taxa present in a community and use a mathematical modeling strategy such as flux balance analysis to predict the metabolic function of the microbial community at a taxon and community-level.[94][95]

As of 2020, understanding is limited due to the missing links between the massive availability of microbiome DNA sequence data on the one hand and limited availability of microbial isolates needed to confirm metagenomic predictions of gene function on the other hand.[1] Metagenome data provides a playground for new predictions, yet much more data is needed to strengthen the links between sequence and rigorous functional predictions. This becomes obvious when considering that the replacement of one single amino acid residue by another may lead to a radical functional change, resulting in an incorrect functional assignment to a given gene sequence.[97] Additionally, cultivation of new strains is needed to help identify the large fraction of unknown sequences obtained from metagenomics analyses, which for poorly studied ecosystems can be more than 70%. Depending on the applied method, even in well-studied microbiomes, 40–70% of the annotated genes in fully sequenced microbial genomes have no known or predicted function.[98] Moreover, current estimates predict that domains with unknown functions will outnumber families of known function very soon.[99] There is a clear need for more classical microbiology including the use of targeted mutants in combination with microbial biochemistry to cope with this challenge. Moreover, there is much more to gain from thorough functional characterization of already discovered protein families with unknown function(s) than from further extending the list of these families.[1] Understanding prokaryotic functional diversity, as of 2019, is challenging as 85 out of the currently established 118 phyla have not had a single species described to this date.[100] [1]
The number of prokaryotic phyla may reach hundreds, and archaeal ones are among the least.[100] The growing gap between the diversity of Bacteria and Archaea held in pure culture and those detected by molecular methods has led to the proposal to establish a formal nomenclature for not-yet cultured taxa, primarily based on sequence information.[101][102] According to this proposal, the concept of Candidatus species would be extended to the groups of closely related genome sequences, and their names would be published following established rules of bacterial nomenclature.[1]
In 1985 Staley and Konopka identified "the great plate count anomaly" which describes the fact that 90 to 99.9% of bacterial species cannot be grown under standard laboratory conditions.[103] For some micro-habitats, especially those with high nutrient content and microbial activity, the proportion of representative strains available in culture relative to the molecular species detected by sequencing grew from 35 to 65%, as it was stated for the gut microbiota.[104] Similar advances are needed for microbial populations from other natural habitats as well as for the eukaryotic members of the microbiome. Micro-eukaryotes, e.g., members of protozoa, fungi, and algae, can often be better cultivated and microscopically studied; however, their phylogeny and taxonomy are more complex and less studied. Interestingly, primer-free 16S and 18S rRNA gene sequencing from various environments has shown that among microeukaryotes there is a huge number of previously not detected taxa.[105][1]
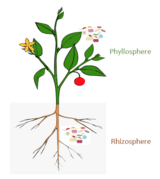
Plant microbiomes

The diagram on the right →
illustrates microbial communities in the soil, air, rhizosphere, phyllosphere, and inside plant tissue (endosphere). In each of these habitats, microbes (represented by colored circles) could interact positively, negatively, or do not interact with other microbes (no lines). Specific microbes, often defined as “hub” or “keystone” species (circles highlighted in bold), are highly connected to other microbes within the networks and likely exert a stronger influence on the structure of microbial communities. (a) Root-associated microbes mainly derive from the soil biome. (b) Leaf-associated microbes originate from various sources such as aerosols, insects, or dust. (c) Relocation between aboveground and belowground microbiota members.[106]
The microbial component of healthy seeds – the seed microbiome – appears to be inherited between plant generations and can dynamically influence germination, plant performance, and survival.[107] As such, methods to optimize the seed microbiomes of major crops could have far-reaching implications for plant breeding and crop improvement to enhance agricultural food, feed, and fiber production.[108]

Marine microbiomes

All animals on Earth form associations with microorganisms, including protists, bacteria, archaea, fungi, and viruses. In the ocean, animal–microbial relationships were historically explored in single host–symbiont systems. However, new explorations into the diversity of microorganisms associating with diverse marine animal hosts is moving the field into studies that address interactions between the animal host and a more multi-member microbiome. The potential for microbiomes to influence the health, physiology, behavior, and ecology of marine animals could alter current understandings of how marine animals adapt to change, and especially the growing climate-related and anthropogenic-induced changes already impacting the ocean environment.[109]
The microbiomes of diverse marine animals are currently under study, from simplistic organisms including sponges[110] and ctenophores [111] to more complex organisms such as sea squirts[112] and sharks.[113][109]
The relationship between the Hawaiian bobtail squid and the bioluminescent bacterium Aliivibrio fischeri is one of the best studied symbiotic relationships in the sea and is a choice system for general symbiosis research. This relationship has provided insight into fundamental processes in animal-microbial symbioses, and especially biochemical interactions and signaling between the host and bacterium.[114][115][109]
The gutless marine oligochaete worm Olavius algarvensis is another relatively well-studied marine host to microbes. These three centimetre long worms reside within shallow marine sediments of the Mediterranean Sea. The worms do not contain a mouth or a digestive or excretory system, but are instead nourished with the help of a suite of extracellular bacterial endosymbionts that reside upon coordinated use of sulfur present in the environment.[116] This system has benefited from some of the most sophisticated 'omics and visualization tools.[117] For example, multi-labeled probing has improved visualization of the microbiome[118] and transcriptomics and proteomics have been applied to examine host–microbiome interactions, including energy transfer between the host and microbes[119] and recognition of the consortia by the worm's innate immune system.[120] The major strength of this system is that it does offer the ability to study host–microbiome interactions with a low diversity microbial consortium, and it also offers a number of host and microbial genomic resources[117][121][109]

Corals are one of the more common examples of an animal host whose symbiosis with microalgae can turn to dysbiosis, and is visibly detected as bleaching. Coral microbiomes have been examined in a variety of studies, which demonstrate how variations in the ocean environment, most notably temperature, light, and inorganic nutrients, affect the abundance and performance of the microalgal symbionts, as well as calcification and physiology of the host.[123][124] Studies have also suggested that resident bacteria, archaea, and fungi additionally contribute to nutrient and organic matter cycling within the coral, with viruses also possibly playing a role in structuring the composition of these members, thus providing one of the first glimpses at a multi-domain marine animal symbiosis.[125] The gammaproteobacterium Endozoicomonas is emerging as a central member of the coral's microbiome, with flexibility in its lifestyle.[122][126] Given the recent mass bleaching occurring on reefs,[127] corals will likely continue to be a useful and popular system for symbiosis and dysbiosis research.[109]
Sponges are common members of the ocean's diverse benthic habitats and their abundance and ability to filter large volumes of seawater have led to the awareness that these organisms play critical roles in influencing benthic and pelagic processes in the ocean.[128] They are one of the oldest lineages of animals, and have a relatively simple body plan that commonly associates with bacteria, archaea, algal protists, fungi, and viruses.[129] Sponge microbiomes are composed of specialists and generalists, and complexity of their microbiome appears to be shaped by host phylogeny.[130] Studies have shown that the sponge microbiome contributes to nitrogen cycling in the oceans, especially through the oxidation of ammonia by archaea and bacteria.[131][132] Most recently, microbial symbionts of tropical sponges were shown to produce and store polyphosphate granules,[133] perhaps enabling the host to survive periods of phosphate depletion in oligotrophic marine environments.[134] The microbiomes of some sponge species do appear to change in community structure in response to changing environmental conditions, including temperature[135] and ocean acidification,[136][137] as well as synergistic impacts.[138]
Underlying complexity

Each microbiome system is suited to address different types of questions based on the culturability of microbes, genetic tractability of microbes and host (where relevant), ability to maintain system in laboratory setting, and ability to make host/environment germfree.[139]
Three different systems are shown in the figure on the right. (A) Pairwise interactions between the soil bacteria Bacillus subtilis and Streptomyces spp. are well-suited for characterizing the functions of secondary metabolites in microbial interactions. (B) The symbiosis between bobtail squid and the marine bacterium Aliivibrio fischeri is fundamental to understanding host and microbial factors that influence colonization. (C) The use of gnotobiotic mice is crucial for making links between host diet and the effects on specific microbial taxa in a community.[139]
Host-microbe coevolution

According to the "separation" approach (upper part of the figure on the right), the microorganisms can be divided into pathogens, neutral, and symbionts, depending on their interaction with their host. The coevolution between host and its associated microbiota may be accordingly described as antagonistic (based on negative interactions) or mutualistic (based on positive interactions).[1][140]
As of 2020, the emergence in publications about opportunistic pathogens and pathobionts has produced a shift towards a holistic approach in the coevolutions theory (lower part of the figure on the right). The holistic approach sees the host and its associated microbiota as one unit (the so-called holobiont), that coevolves as one entity. According to the holistic approach, holobiont's disease state is linked to dysbiosis, low diversity of the associated microbiota, and their variability: a so-called pathobiome state. The healthy state, on the other hand, is accompanied with eubiosis, high diversity, and uniformity of the respective microbiota.[1]
See also
- Earth Microbiome Project
- Human microbiome
- Initial acquisition of microbiota
- Microbial population biology
- Microbiomes of the built environment
- Mycobiome
References
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap Berg, Gabriele; Rybakova, Daria; Fischer, Doreen; Cernava, Tomislav; Vergès, Marie-Christine Champomier; Charles, Trevor; Chen, Xiaoyulong; Cocolin, Luca; Eversole, Kellye; Corral, Gema Herrero; Kazou, Maria; Kinkel, Linda; Lange, Lene; Lima, Nelson; Loy, Alexander; MacKlin, James A.; Maguin, Emmanuelle; Mauchline, Tim; McClure, Ryan; Mitter, Birgit; Ryan, Matthew; Sarand, Inga; Smidt, Hauke; Schelkle, Bettina; Roume, Hugo; Kiran, G. Seghal; Selvin, Joseph; Souza, Rafael Soares Correa de; Van Overbeek, Leo; et al. (2020). "Microbiome definition re-visited: Old concepts and new challenges". Microbiome. 8 (1): 103. doi:10.1186/s40168-020-00875-0. PMC 7329523. PMID 32605663.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ "BioConcepts". www.biological-concepts.com. Retrieved 18 December 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ "microbiome". Oxford English Dictionary (Online ed.). Oxford University Press. Retrieved 18 December 2020. (Subscription or participating institution membership required.)
- ^ a b c d e Whipps J., Lewis K. and Cooke R. (1988) "Mycoparasitism and plant disease control". In: Burge M (Ed.) Fungi in Biological Control Systems, Manchester University Press, pages 161–187. ISBN 9780719019791.
- ^ Hiltner L. (1902) "Die Keimungsverhältnisse der Leguminosensamen und ihre Beeinflussung durch Organismenwirkung". In: Parey P and Springer J (Eds.) Arb Biol Abt Land u Forstw K Gsndhtsamt, 3, Berlin. Pages 1-545.
- ^ Metchnikoff E. The prolongation of life: optimistic studies. GP Putnam's Sons; 1908.
- ^ a b Bassler, B.L. (2002) "Small talk: cell-to-cell communication in bacteria". Cell, 109(4): 421–424. doi:10.1016/S0092-8674(02)00749-3.
- ^ Brul, S., Kallemeijn, W. and Smits, G. (2008) "Functional genomics for food microbiology: molecular mechanisms of weak organic-acid preservative adaptation in yeast". CAB Rev, 3: 1–14. doi:10.1079/PAVSNNR20083005.
- ^ Woese, C.R. and Fox, G.E. (1977) "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences, 74(11): 5088–5090. doi:10.1073/pnas.74.11.5088.
- ^ Uksa, M., Schloter, M., Endesfelder, D., Kublik, S., Engel, M., Kautz, T., Köpke, U. and Fischer, D. (2015) "Prokaryotes in subsoil—evidence for a strong spatial separation of different phyla by analysing co-occurrence networks". Frontiers in microbiology, 6: 1269. doi:10.3389/fmicb.2015.01269.
- ^ Maritz, J.M., Rogers, K.H., Rock, T.M., Liu, N., Joseph, S., Land, K.M. and Carlton, J.M. (2017) "An 18S rRNA workflow for characterizing protists in sewage, with a focus on zoonotic trichomonads". Microbial ecology, 74(4): 923–936. doi:10.1007/s00248-017-0996-9.
- ^ Purahong, W., Wubet, T., Lentendu, G., Schloter, M., Pecyna, M.J., Kapturska, D., Hofrichter, M., Krüger, D. and Buscot, F. (2016) "Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition". Molecular Ecology, 25(16): 4059–4074. doi:10.1111/mec.13739.
- ^ Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K. and Knight, R. (2012) "Diversity, stability and resilience of the human gut microbiota". Nature, 489(7415): 220–230. doi:10.1038/nature11550.
- ^ Venter, J.C., Remington, K., Heidelberg, J.F., Halpern, A.L., Rusch, D., Eisen, J.A., Wu, D., Paulsen, I., Nelson, K.E., Nelson, W. and Fouts, D.E. (2004) "Environmental genome shotgun sequencing of the Sargasso Sea". Science, 304(5667): 66–74. doi:10.1126/science.1093857.
- ^ Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L. and Law, M. (2012) "Comparison of next-generation sequencing systems". BioMed Research International, 2012: 251364. doi:10.1155/2012/251364.
- ^ Stegen, J.C., Bottos, E.M. and Jansson, J.K. (2018) "A unified conceptual framework for prediction and control of microbiomes". Current Opinion in Microbiology, 44: 20–27. doi:10.1016/j.mib.2018.06.002.
- ^ Knight, R., Vrbanac, A., Taylor, B.C., Aksenov, A., Callewaert, C., Debelius, J., Gonzalez, A., Kosciolek, T., McCall, L.I., McDonald, D. and Melnik, A.V. (2018) "Best practices for analysing microbiomes". Nature Reviews Microbiology, 16(7): 410–422. doi:10.1038/s41579-018-0029-9.
- ^ Lane, Nick (2015). "The unseen world: Reflections on Leeuwenhoek (1677) 'Concerning little animals'". Philosophical Transactions of the Royal Society B: Biological Sciences. 370 (1666). doi:10.1098/rstb.2014.0344. PMC 4360124. PMID 25750239.
- ^ Jarvis, Charles E. (2016). "Pier Antonio Micheli (1679–1737) and Carl Linnaeus (1707–1778)". Webbia. 71: 1–24. doi:10.1080/00837792.2016.1147210. S2CID 88308313.
- ^ Riedel, Stefan (2005). "Edward Jenner and the History of Smallpox and Vaccination". Baylor University Medical Center Proceedings. 18 (1): 21–25. doi:10.1080/08998280.2005.11928028. PMC 1200696. PMID 16200144.
- ^ Martini, Alessandro (1993). "Origin and domestication of the wine yeast Saccharomyces cerevisiae". Journal of Wine Research. 4 (3): 165–176. doi:10.1080/09571269308717966.
- ^ Berche, P. (2012). "Louis Pasteur, from crystals of life to vaccination". Clinical Microbiology and Infection. 18. Elsevier BV: 1–6. doi:10.1111/j.1469-0691.2012.03945.x. ISSN 1198-743X.
- ^ Evans, A.S. (1976) "Causation and disease: the Henle-Koch postulates revisited. The Yale journal of biology and medicine, 49(2): 175.
- ^ Dworkin, Martin; Gutnick, David (2012). "Sergei Winogradsky: A founder of modern microbiology and the first microbial ecologist". FEMS Microbiology Reviews. 36 (2): 364–379. doi:10.1111/j.1574-6976.2011.00299.x. PMID 22092289.
- ^ Hartmann, Anton; Rothballer, Michael; Schmid, Michael (2008). "Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research". Plant and Soil. 312 (1–2): 7–14. doi:10.1007/s11104-007-9514-z. S2CID 4419735.
- ^ Hartmann, Anton; Rothballer, Michael; Schmid, Michael (2008). "Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research". Plant and Soil. 312 (1–2): 7–14. doi:10.1007/s11104-007-9514-z. S2CID 4419735.
- ^ "The Fluorescence Microscope". Microscopes—Help Scientists Explore Hidden Worlds. The Nobel Foundation. Retrieved 28 September 2008.
- ^ Borman, S., Russell, H. and Siuzdak, G., (2003) "A Mass Spec Timeline Developing techniques to measure mass has been a Nobel pursuit. Todays Chemist at Work, 12(9): 47–50.
- ^ Waksman, Selman A. (1953). "Sergei Nikolaevitch Winogradsky: 1856-1953". Science. 118 (3054): 36–37. Bibcode:1953Sci...118...36W. doi:10.1126/science.118.3054.36. PMID 13076173.
- ^ Griffith, Fred (1928). "The Significance of Pneumococcal Types". Journal of Hygiene. 27 (2): 113–159. doi:10.1017/S0022172400031879. PMC 2167760. PMID 20474956.
- ^ Hayes, W. (1966) "Genetic Transformation: a Retrospective Appreciation", First Griffith Memorial Lecture. Microbiology, 45(3): 385-397.
- ^ American Chemical Society (1999) Discovery and Development of Penicillin, 1928–1945. International Historic Chemical Landmarks, The Alexander Fleming Laboratory Museum, London.
- ^ Ruska, Ernst (1987). "The Development of the Electron Microscope and of Electron Microscopy(Nobel Lecture)". Angewandte Chemie International Edition in English. 26 (7): 595–605. doi:10.1002/anie.198705953.
- ^ Avery, O. T.; MacLeod, C. M.; McCarty, M. (1979). "Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III". Journal of Experimental Medicine. 149 (2): 297–326. doi:10.1084/jem.149.2.297. PMC 2184805. PMID 33226.
- ^ o'Malley, Maureen A. (2018). "The Experimental Study of Bacterial Evolution and Its Implications for the Modern Synthesis of Evolutionary Biology". Journal of the History of Biology. 51 (2): 319–354. doi:10.1007/s10739-017-9493-8. PMID 28980196. S2CID 4055566.
- ^ Rich, Alexander (2003). "The double helix: A tale of two puckers". Nature Structural & Molecular Biology. 10 (4): 247–249. doi:10.1038/nsb0403-247. PMID 12660721. S2CID 6089989.
- ^ a b Cassidy, Andrew; Jones, Julia (2014). "Developments in in situ hybridisation". Methods. 70 (1): 39–45. doi:10.1016/j.ymeth.2014.04.006. PMID 24747923.
- ^ Crick, Francis (1970). "Central Dogma of Molecular Biology". Nature. 227 (5258): 561–563. Bibcode:1970Natur.227..561C. doi:10.1038/227561a0. PMID 4913914. S2CID 4164029.
- ^ Meyer, Veronika (2013). Practical high-performance liquid chromatography. Hoboken, N.J: Wiley. ISBN 978-1-118-68134-3. OCLC 864917338.
- ^ Grunstein, M.; Hogness, D. S. (1975). "Colony hybridization: A method for the isolation of cloned DNAs that contain a specific gene". Proceedings of the National Academy of Sciences. 72 (10): 3961–3965. Bibcode:1975PNAS...72.3961G. doi:10.1073/pnas.72.10.3961. PMC 433117. PMID 1105573.
- ^ Sanger, F.; Nicklen, S.; Coulson, A. R. (1977). "DNA sequencing with chain-terminating inhibitors". Proceedings of the National Academy of Sciences. 74 (12): 5463–5467. Bibcode:1977PNAS...74.5463S. doi:10.1073/pnas.74.12.5463. PMC 431765. PMID 271968.
- ^ Heather, James M.; Chain, Benjamin (2016). "The sequence of sequencers: The history of sequencing DNA". Genomics. 107: 1–8. doi:10.1016/j.ygeno.2015.11.003. PMID 26554401. S2CID 27846422.
- ^ Woese, C. R.; Fox, G. E. (1977). "Phylogenetic structure of the prokaryotic domain: The primary kingdoms". Proceedings of the National Academy of Sciences. 74 (11): 5088–5090. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. PMC 432104. PMID 270744.
- ^ Eme, Laura; Spang, Anja; Lombard, Jonathan; Stairs, Courtney W.; Ettema, Thijs J. G. (2017). "Archaea and the origin of eukaryotes". Nature Reviews Microbiology. 15 (12): 711–723. doi:10.1038/nrmicro.2017.133. PMID 29123225. S2CID 8666687.
- ^ Fiers, W.; Contreras, R.; Duerinck, F.; Haegeman, G.; Iserentant, D.; Merregaert, J.; Min Jou, W.; Molemans, F.; Raeymaekers, A.; Van Den Berghe, A.; Volckaert, G.; Ysebaert, M. (1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–507. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203. S2CID 4289674.
- ^ Prusiner, Stanley B. (1982). "Novel Proteinaceous Infectious Particles Cause Scrapie". Science. 216 (4542): 136–144. Bibcode:1982Sci...216..136P. doi:10.1126/science.6801762. PMID 6801762.
- ^ Mullis, K.B. (1990) "The unusual origin of the polymerase chain reaction". Scientific American, 262(4): 56–65.
- ^ Higuchi, Russell; Fockler, Carita; Dollinger, Gavin; Watson, Robert (1993). "Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions". Nature Biotechnology. 11 (9): 1026–1030. doi:10.1038/nbt0993-1026. PMID 7764001. S2CID 5714001.
- ^ Bentleylawrence, J.; Villnave, C. A.; Singer, R. H. (1988). "Sensitive, high-resolution chromatin and chromosome mapping in situ: Presence and orientation of two closely integrated copies of EBV in a lymphoma line". Cell. 52 (1): 51–61. doi:10.1016/0092-8674(88)90530-2. PMID 2830981. S2CID 17565963.
- ^ Margulis, Lynn (1991). Symbiosis as a source of evolutionary innovation : speciation and morphogenesis. Cambridge, Mass: MIT Press. ISBN 978-0-262-13269-5. OCLC 22597587.
- ^ Flemming, Hans-Curt (1993). "Biofilms and Environmental Protection". Water Science and Technology. 27 (7–8): 1–10. doi:10.2166/wst.1993.0528.
- ^ Flemming (2011). Biofilm highlights. Heidelberg New York: Springer-Verlag Berlin Heidelberg. ISBN 978-3-642-19940-0. OCLC 769756150.
- ^ Amann, R. I.; Ludwig, W.; Schleifer, K. H. (1995). "Phylogenetic identification and in situ detection of individual microbial cells without cultivation". Microbiological Reviews. 59 (1): 143–169. doi:10.1128/mr.59.1.143-169.1995. PMC 239358. PMID 7535888.
- ^ Fleischmann, Robert D.; et al. (1995). "Whole-Genome Random Sequencing and Assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
- ^ Kulski, Jerzy K. (2016). "Next-Generation Sequencing — an Overview of the History, Tools, and "Omic" Applications". Next Generation Sequencing - Advances, Applications and Challenges. doi:10.5772/61964. ISBN 978-953-51-2240-1.
- ^ Stern, A.; Mick, E.; Tirosh, I.; Sagy, O.; Sorek, R. (2012). "CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome". Genome Research. 22 (10): 1985–1994. doi:10.1101/gr.138297.112. PMC 3460193. PMID 22732228.
- ^ Schadt, E. E.; Turner, S.; Kasarskis, A. (2010). "A window into third-generation sequencing". Human Molecular Genetics. 19 (R2): R227–R240. doi:10.1093/hmg/ddq416. PMID 20858600.
- ^ Vogel, Timothy M.; Simonet, Pascal; Jansson, Janet K.; Hirsch, Penny R.; Tiedje, James M.; Van Elsas, Jan Dirk; Bailey, Mark J.; Nalin, Renaud; Philippot, Laurent (2009). "Terra Genome: A consortium for the sequencing of a soil metagenome". Nature Reviews Microbiology. 7 (4): 252. doi:10.1038/nrmicro2119. S2CID 2144462.
- ^ Gilbert, Jack A.; Meyer, Folker; Jansson, Janet; Gordon, Jeff; Pace, Norman; Tiedje, James; Ley, Ruth; Fierer, Noah; Field, Dawn; Kyrpides, Nikos; Glöckner, Frank-Oliver; Klenk, Hans-Peter; Wommack, K. Eric; Glass, Elizabeth; Docherty, Kathryn; Gallery, Rachel; Stevens, Rick; Knight, Rob (2010). "The Earth Microbiome Project: Meeting report of the "1st EMP meeting on sample selection and acquisition" at Argonne National Laboratory October 6th 2010". Standards in Genomic Sciences. 3 (3): 249–253. doi:10.4056/aigs.1443528. PMC 3035312. PMID 21304728.
- ^ Konopka, A. (2009) "What is microbial community ecology?" The ISME Journal, 3(11): 1223–1230. {{doi|Konopka, A., 2009. What is microbial community ecology?. The ISME journal, 3(11), pp.1223-1230. doi:10.1038/ismej.2009.88.
- ^ a b Lederberg, J. and McCray, A.T. (2001) "'Ome Sweet'Omics--A genealogical treasury of words". The Scientist, 15(7): 8.
- ^ a b c d e Marchesi, J.R. and Ravel, J. (2015) "The vocabulary of microbiome research: a proposal". Microbiome, 3(31). doi:10.1186/s40168-015-0094-5.
- ^ Prosser, J.I., Bohannan, B.J., Curtis, T.P., Ellis, R.J., Firestone, M.K., Freckleton, R.P., Green, J.L., Green, L.E., Killham, K., Lennon, J.J. and Osborn, A.M. (2007) "The role of ecological theory in microbial ecology". Nature Reviews Microbiology, 5(5): 384–392. doi:10.1038/nrmicro1643.
- ^ del Carmen Orozco-Mosqueda, M., del Carmen Rocha-Granados, M., Glick, B.R. and Santoyo, G. (2018) "Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms". Microbiological Research, 208: 25–31. doi:10.1016/j.micres.2018.01.005.
- ^ a b Merriam-Webster Dictionary – microbiome.
- ^ Human Microbiome Project. Accessed 25 Aug 2020.
- ^ Nature.com: Microbiome. Accessed 25 August 2020.
- ^ ScienceDirect: Microbiome Accessed 25 August 2020.
- ^ Arevalo, P., VanInsberghe, D., Elsherbini, J., Gore, J. and Polz, M.F. (2019) "A reverse ecology approach based on a biological definition of microbial populations". Cell, 178(4): 820–834. doi:10.1016/j.cell.2019.06.033.
- ^ Schlaeppi, K. and Bulgarelli, D. (2015) "The plant microbiome at work". Molecular Plant-Microbe Interactions, 28(3): 212–217. doi:10.1094/MPMI-10-14-0334-FI.
- ^ Rogers Y-H and Zhang C. (2016) "Genomic Technologies in Medicine and Health: Past, Present, and Future". In: Kumar D and Antonarakis S. (Eds.) Medical and Health Genomics. Oxford: Academic Press, pages 15–28. ISBN 9780127999227.
- ^ Ho, H.E. and Bunyavanich, S. (2018) "Role of the microbiome in food allergy". Current allergy and asthma reports, 18(4): 27. doi:10.1007/s11882-018-0780-z.
- ^ Whiteside, S.A., Razvi, H., Dave, S., Reid, G. and Burton and J.P. (2015) "The microbiome of the urinary tract—a role beyond infection". Nature Reviews Urology, 12(2): 81–90. doi:10.1038/nrurol.2014.361.
- ^ MicrobiomeSupport project
- ^ Carini, Paul (2016) A census of the dead: the story behind microbial ‘relic DNA’ in soil Nature Research: Microbiology.
- ^ Carini, P., Marsden, P.J., Leff, J.W., Morgan, E.E., Strickland, M.S. and Fierer, N. (2016) "Relic DNA is abundant in soil and obscures estimates of soil microbial diversity". Nature Microbiology, 2(3): 1–6. doi:10.1038/nmicrobiol.2016.242.
- ^ a b Lennon, J.T., Muscarella, M.E., Placella, S.A. and Lehmkuhl, B.K. (2018) "How, when, and where relic DNA affects microbial diversity". mBio, 9(3). doi:10.1128/mBio.00637-18.
- ^ Dupré JO, O’Malley MA (2009) "Varieties of living things: life at the intersection of lineage and metabolism". In: Normandin S and Wolfe C (Eds.) Vitalism and the Scientific Image in Post-Enlightenment Life Science 1800–2010. Dordrecht: Springer, pages 311–344. ISBN 9789400724457.
- ^ McDaniel, L., Breitbart, M., Mobberley, J., Long, A., Haynes, M., Rohwer, F. and Paul, J.H., 2008. Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One, 3(9), p.e3263. doi:10.1371/journal.pone.0003263.
- ^ Banerjee, Samiran; Schlaeppi, Klaus; Van Der Heijden, Marcel G. A. (2018). "Keystone taxa as drivers of microbiome structure and functioning" (PDF). Nature Reviews Microbiology. 16 (9): 567–576. doi:10.1038/s41579-018-0024-1. PMID 29789680. S2CID 46895123.
- ^ Ho, Adrian; Lonardo, D. Paolo Di; Bodelier, Paul L. E. (2017). "Revisiting life strategy concepts in environmental microbial ecology". FEMS Microbiology Ecology. 93 (3): fix006. doi:10.1093/femsec/fix006. PMID 28115400.
- ^ Banerjee, Samiran; Schlaeppi, Klaus; Van Der Heijden, Marcel G. A. (2018). "Keystone taxa as drivers of microbiome structure and functioning" (PDF). Nature Reviews Microbiology. 16 (9): 567–576. doi:10.1038/s41579-018-0024-1. PMID 29789680. S2CID 46895123.
- ^ Papenfort, Kai; Bassler, Bonnie L. (2016). "Quorum sensing signal–response systems in Gram-negative bacteria". Nature Reviews Microbiology. 14 (9): 576–588. doi:10.1038/nrmicro.2016.89. PMC 5056591. PMID 27510864.
- ^ Lovley, Derek R. (2017). "Syntrophy Goes Electric: Direct Interspecies Electron Transfer". Annual Review of Microbiology. 71: 643–664. doi:10.1146/annurev-micro-030117-020420. PMID 28697668.
- ^ Schmidt, Ruth; Etalo, Desalegn W.; De Jager, Victor; Gerards, Saskia; Zweers, Hans; De Boer, Wietse; Garbeva, Paolina (2016). "Microbial Small Talk: Volatiles in Fungal–Bacterial Interactions". Frontiers in Microbiology. 6: 1495. doi:10.3389/fmicb.2015.01495. PMC 4700264. PMID 26779150.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Zhang, Yuanchen; Kastman, Erik K.; Guasto, Jeffrey S.; Wolfe, Benjamin E. (2018). "Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes". Nature Communications. 9 (1): 336. Bibcode:2018NatCo...9..336Z. doi:10.1038/s41467-017-02522-z. PMC 5780524. PMID 29362365.
- ^ Worrich, Anja; Stryhanyuk, Hryhoriy; Musat, Niculina; König, Sara; Banitz, Thomas; Centler, Florian; Frank, Karin; Thullner, Martin; Harms, Hauke; Richnow, Hans-Hermann; Miltner, Anja; Kästner, Matthias; Wick, Lukas Y. (2017). "Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments". Nature Communications. 8: 15472. Bibcode:2017NatCo...815472W. doi:10.1038/ncomms15472. PMC 5467244. PMID 28589950.
- ^ Chevrette, Marc G.; Bratburd, Jennifer R.; Currie, Cameron R.; Stubbendieck, Reed M. (2019). "Experimental Microbiomes: Models Not to Scale". mSystems. 4 (4). doi:10.1128/mSystems.00175-19. PMC 6667727. PMID 31363014.
- ^ Böttcher, Thomas (2020). "Unveiling the Hidden Theatre of Microbes". ChemBioChem. 21 (20): 2869. doi:10.1002/cbic.202000554. PMID 32869434. S2CID 221403702.
- ^ Anantharaman, Karthik; Brown, Christopher T.; Hug, Laura A.; Sharon, Itai; Castelle, Cindy J.; Probst, Alexander J.; Thomas, Brian C.; Singh, Andrea; Wilkins, Michael J.; Karaoz, Ulas; Brodie, Eoin L.; Williams, Kenneth H.; Hubbard, Susan S.; Banfield, Jillian F. (2016). "Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system". Nature Communications. 7: 13219. Bibcode:2016NatCo...713219A. doi:10.1038/ncomms13219. PMC 5079060. PMID 27774985.
- ^ Pasolli, Edoardo; Asnicar, Francesco; Manara, Serena; Zolfo, Moreno; Karcher, Nicolai; Armanini, Federica; Beghini, Francesco; Manghi, Paolo; Tett, Adrian; Ghensi, Paolo; Collado, Maria Carmen; Rice, Benjamin L.; Dulong, Casey; Morgan, Xochitl C.; Golden, Christopher D.; Quince, Christopher; Huttenhower, Curtis; Segata, Nicola (2019). "Extensive Unexplored Human Microbiome Diversity Revealed by over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle". Cell. 176 (3): 649–662.e20. doi:10.1016/j.cell.2019.01.001. PMC 6349461. PMID 30661755.
- ^ Kumar, Manish; Ji, Boyang; Zengler, Karsten; Nielsen, Jens (23 July 2019). "Modelling approaches for studying the microbiome". Nature Microbiology. 4 (8): 1253–1267. doi:10.1038/s41564-019-0491-9. ISSN 2058-5276.
- ^ Borenstein, Elhanan (15 May 2012). "Computational systems biology and in silico modeling of the human microbiome". Briefings in Bioinformatics. 13: 769–780.
- ^ Colarusso, Analeigha V.; Goodchild-Michelman, Isabella; Rayle, Maya; Zomorrodi, Ali R. (4 June 2021). "Computational modeling of metabolism in microbial communities on a genome-scale". Current Opinion in Systems Biology. 26: 46–57. doi:10.1016/j.coisb.2021.04.001. ISSN 2452-3100.
- ^ Biggs, Matthew B.; Medlock, Gregory L.; Kolling, Glynis L.; Papin, Jason A. (24 June 2015). "Metabolic network modeling of microbial communities". Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 7 (5): 317–334. doi:10.1002/wsbm.1308. ISSN 1939-5094. PMC 4575871.
- ^ Hugenholtz, P. (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biology. 3 (2): reviews0003.reviews0001. doi:10.1186/gb-2002-3-2-reviews0003. PMC 139013. PMID 11864374.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Bloom, J. D.; Arnold, F. H. (2009). "In the light of directed evolution: Pathways of adaptive protein evolution". Proceedings of the National Academy of Sciences. 106: 9995–10000. doi:10.1073/pnas.0901522106. PMC 2702793. PMID 19528653.
- ^ Heintz-Buschart, Anna; Wilmes, Paul (2018). "Human Gut Microbiome: Function Matters". Trends in Microbiology. 26 (7): 563–574. doi:10.1016/j.tim.2017.11.002. PMID 29173869.
- ^ Baric, Ralph S.; Crosson, Sean; Damania, Blossom; Miller, Samuel I.; Rubin, Eric J. (2016). "Next-Generation High-Throughput Functional Annotation of Microbial Genomes". mBio. 7 (5). doi:10.1128/mBio.01245-16. PMC 5050336. PMID 27703071.
- ^ a b Overmann, Jörg; Huang, Sixing; Nübel, Ulrich; Hahnke, Richard L.; Tindall, Brian J. (2019). "Relevance of phenotypic information for the taxonomy of not-yet-cultured microorganisms". Systematic and Applied Microbiology. 42 (1): 22–29. doi:10.1016/j.syapm.2018.08.009. PMID 30197212.
- ^ Konstantinidis, Konstantinos T.; Rosselló-Móra, Ramon; Amann, Rudolf (2017). "Uncultivated microbes in need of their own taxonomy". The ISME Journal. 11 (11): 2399–2406. doi:10.1038/ismej.2017.113. PMC 5649169. PMID 28731467.
- ^ Chuvochina, Maria; Rinke, Christian; Parks, Donovan H.; Rappé, Michael S.; Tyson, Gene W.; Yilmaz, Pelin; Whitman, William B.; Hugenholtz, Philip (2019). "The importance of designating type material for uncultured taxa". Systematic and Applied Microbiology. 42 (1): 15–21. doi:10.1016/j.syapm.2018.07.003. PMID 30098831.
- ^ Staley, J. T.; Konopka, A. (1985). "Measurement of in Situ Activities of Nonphotosynthetic Microorganisms in Aquatic and Terrestrial Habitats". Annual Review of Microbiology. 39: 321–346. doi:10.1146/annurev.mi.39.100185.001541. PMID 3904603.
- ^ Lagkouvardos, Ilias; Overmann, Jörg; Clavel, Thomas (2017). "Cultured microbes represent a substantial fraction of the human and mouse gut microbiota". Gut Microbes. 8 (5): 493–503. doi:10.1080/19490976.2017.1320468. PMC 5628658. PMID 28418756.
- ^ Karst, Søren M.; Dueholm, Morten S.; McIlroy, Simon J.; Kirkegaard, Rasmus H.; Nielsen, Per H.; Albertsen, Mads (2018). "Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias". Nature Biotechnology. 36 (2): 190–195. doi:10.1038/nbt.4045. PMID 29291348. S2CID 205285663.
- ^ a b Hassani, M.A., Durán, P. and Hacquard, S. (2018) "Microbial interactions within the plant holobiont". Microbiome, 6(1): 58. doi:10.1186/s40168-018-0445-0.
 Material was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License
Material was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License
- ^ Matsumoto, Haruna; Fan, Xiaoyan; Wang, Yue; Kusstatscher, Peter; Duan, Jie; Wu, Sanling; Chen, Sunlu; Qiao, Kun; Wang, Yiling; Ma, Bin; Zhu, Guonian (2021). "Bacterial seed endophyte shapes disease resistance in rice". Nature Plants. 7 (1): 60–72. doi:10.1038/s41477-020-00826-5. ISSN 2055-0278.
- ^ a b Mitter, B., Pfaffenbichler, N., Flavell, R., Compant, S., Antonielli, L., Petric, A., Berninger, T., Naveed, M., Sheibani-Tezerji, R., von Maltzahn, G. and Sessitsch, A. (2017) "A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds". Frontiers in Microbiology, 8: 11. doi:10.3389/fmicb.2017.00011.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d e f Apprill, A. (2017) "Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean". Frontiers in Marine Science, 4: 222. doi:10.3389/fmars.2017.00222.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Webster, N.S., Negri, A.P., Botté, E.S., Laffy, P.W., Flores, F., Noonan, S., Schmidt, C. and Uthicke, S. (2016) "Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification". Scientific reports, 6: 19324. doi:10.1038/srep19324.
- ^ Daniels, C. and Breitbart, M. (2012) "Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovata". FEMS Microbiology Ecology, 82(1): 90–101. doi:10.1111/j.1574-6941.2012.01409.x.
- ^ Blasiak, L.C., Zinder, S.H., Buckley, D.H. and Hill, R.T. (2014) "Bacterial diversity associated with the tunic of the model chordate Ciona intestinalis". The ISME Journal, 8(2): 309–320. doi:10.1038/ismej.2013.156.
- ^ Givens, C.E., Ransom, B., Bano, N. and Hollibaugh, J.T. (2015) "Comparison of the gut microbiomes of 12 bony fish and 3 shark species". Marine Ecology Progress Series, 518: 209–223. doi:10.3354/meps11034.
- ^ McFall-Ngai, M.J. (2000) "Negotiations between animals and bacteria: the ‘diplomacy’of the squid-vibrio symbiosis". Comparative Biochemistry and Physiology, Part A: Molecular & Integrative Physiology, 126(4): 471–480. doi:10.1016/S1095-6433(00)00233-6.
- ^ McFall-Ngai, M. (2014) "Divining the essence of symbiosis: insights from the squid-vibrio model". PLoS Biology, 12(2): e1001783. doi:10.1371/journal.pbio.1001783.
- ^ Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., Wagner, M., Erséus, C., Thiermann, F., Krieger, J. and Giere, O. (2001) "Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm". Nature, 411(6835): 298–302. doi:10.1038/35077067.
- ^ a b Woyke, T., Teeling, H., Ivanova, N.N., Huntemann, M., Richter, M., Gloeckner, F.O., Boffelli, D., Anderson, I.J., Barry, K.W., Shapiro, H.J. and Szeto, E. (2006) "Symbiosis insights through metagenomic analysis of a microbial consortium". Nature, 443(7114): 950–955. doi:10.1038/nature05192.
- ^ Schimak, M.P., Kleiner, M., Wetzel, S., Liebeke, M., Dubilier, N. and Fuchs, B.M. (2016) "MiL-FISH: Multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells". Applied and Environmental Microbiology, 82(1): 62–70. doi:10.1128/AEM.02776-15.
- ^ Kleiner, M., Wentrup, C., Lott, C., Teeling, H., Wetzel, S., Young, J., Chang, Y.J., Shah, M., VerBerkmoes, N.C., Zarzycki, J. and Fuchs, G. (2012) "Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use". Proceedings of the National Academy of Sciences, 109(19): E1173–E1182. doi:10.1073/pnas.1121198109.
- ^ Wippler, J., Kleiner, M., Lott, C., Gruhl, A., Abraham, P.E., Giannone, R.J., Young, J.C., Hettich, R.L. and Dubilier, N. (2016) "Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis". BMC Genomics, 17(1): 942. doi:10.1186/s12864-016-3293-y.
- ^ Ruehland, C., Blazejak, A., Lott, C., Loy, A., Erséus, C. and Dubilier, N. (2008) "Multiple bacterial symbionts in two species of co‐occurring gutless oligochaete worms from Mediterranean sea grass sediments". Environmental microbiology, 10(12): 3404–3416. doi:10.1111/j.1462-2920.2008.01728.x.
- ^ a b Neave, M.J., Apprill, A., Ferrier-Pagès, C. and Voolstra, C.R. (2016) "Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas". Applied Microbiology and Biotechnology, 100(19): 8315–8324. doi:10.1007/s00253-016-7777-0.
- ^ Dubinsky, Z. and Jokiel, P.L. (1994) "Ratio of energy and nutrient fluxes regulates symbiosis between zooxanthellae and corals". Pacific Science, 48(3): 313–324.
- ^ Anthony, K.R., Kline, D.I., Diaz-Pulido, G., Dove, S. and Hoegh-Guldberg, O.(2008) "Ocean acidification causes bleaching and productivity loss in coral reef builders". Proceedings of the National Academy of Sciences, 105(45): 17442–17446. doi:10.1073/pnas.0804478105.
- ^ Bourne, D.G., Morrow, K.M. and Webster, N.S. (2016) "Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems". Annual Review of Microbiology, 70: 317–340. doi:10.1146/annurev-micro-102215-095440.
- ^ Neave, M.J., Michell, C.T., Apprill, A. and Voolstra, C.R. (2017) "Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts". Scientific Reports, 7: 40579. doi:10.1038/srep40579.
- ^ Hughes, T.P., Kerry, J.T., Álvarez-Noriega, M., Álvarez-Romero, J.G., Anderson, K.D., Baird, A.H., Babcock, R.C., Beger, M., Bellwood, D.R., Berkelmans, R. and Bridge, T.C. (2017) "Global warming and recurrent mass bleaching of corals". Nature, 543(7645): 373–377. doi:10.1038/nature21707.
- ^ Bell, J.J. (2008) "The functional roles of marine sponges". Estuarine, Coastal and Shelf Science, 79(3): 341–353. doi:10.1016/j.ecss.2008.05.002.
- ^ Webster, N.S. and Thomas, T. (2016) "The sponge hologenome". mBio, 7(2). doi:10.1128/mBio.00135-16.
- ^ Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J.R., Easson, C., Astudillo-García, C., Olson, J.B., Erwin, P.M., López-Legentil, S., Luter, H. and Chaves-Fonnegra, A. (2016) "Diversity, structure and convergent evolution of the global sponge microbiome". Nature Communications, 7(1): 1-12. doi:10.1038/ncomms11870.
- ^ Bayer, K., Schmitt, S. and Hentschel, U. (2008) "Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba". Environmental Microbiology, 10(11): 2942–2955. doi:10.1111/j.1462-2920.2008.01582.x.
- ^ Radax, R., Hoffmann, F., Rapp, H.T., Leininger, S. and Schleper, C. (2012) "Ammonia‐oxidizing archaea as main drivers of nitrification in cold‐water sponges". Environmental Microbiology, 14(4): 909_923. doi:10.1111/j.1462-2920.2011.02661.x.
- ^ Zhang, F., Blasiak, L.C., Karolin, J.O., Powell, R.J., Geddes, C.D. and Hill, R.T. (2015) "Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges". Proceedings of the National Academy of Sciences, 112(14): 4381–4386. doi:10.1073/pnas.1423768112.
- ^ Colman, A.S. (2015) "Sponge symbionts and the marine P cycle". Proceedings of the National Academy of Sciences, 112(14): 4191–4192. doi:10.1073/pnas.1502763112.
- ^ Simister, R., Taylor, M.W., Tsai, P., Fan, L., Bruxner, T.J., Crowe, M.L. and Webster, N. (2012) "Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environmental Microbiology, 14(12): 3232–3246. doi:10.1111/1462-2920.12010.
- ^ Morrow, K.M., Bourne, D.G., Humphrey, C., Botté, E.S., Laffy, P., Zaneveld, J., Uthicke, S., Fabricius, K.E. and Webster, N.S. (2015) "Natural volcanic CO 2 seeps reveal future trajectories for host–microbial associations in corals and sponges". The ISME Journal, 9(4): 894–908. doi:10.1038/ismej.2014.188.
- ^ Ribes, M., Calvo, E., Movilla, J., Logares, R., Coma, R. and Pelejero, C. (2016) "Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environmental Microbiology Reports, 8(4): 536–544. doi:10.1111/1758-2229.12430.
- ^ Lesser, M.P., Fiore, C., Slattery, M. and Zaneveld, J. (2016) "Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta". Journal of Experimental Marine Biology and Ecology, 475: 11–18. doi:10.1016/j.jembe.2015.11.004.
- ^ a b c Chevrette, M.G., Bratburd, J.R., Currie, C.R. and Stubbendieck, R.M. (2019 "Experimental Microbiomes: Models Not to Scale". mSystems, 4(4): e00175-19. doi:10.1128/mSystems.00175-19.
- ^ Leftwich, Philip T.; Edgington, Matthew P.; Chapman, Tracey (9 September 2020). "Transmission efficiency drives host–microbe associations". Proceedings of the Royal Society B: Biological Sciences. 287 (1934): 20200820. doi:10.1098/rspb.2020.0820. ISSN 0962-8452. PMC 7542779. PMID 32873208.

