Acinetobacter
| Acinetobacter | |
|---|---|
| |
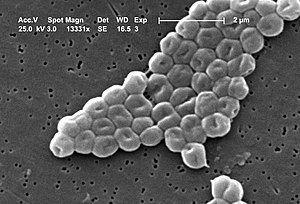
| Acinetobacter baumannii | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Acinetobacter Brisou & Prévot 1954
|
Acinetobacter [asz−in−ée−toe–back−ter] is a genus of Gram-negative bacteria belonging to the Gammaproteobacteria. Acinetobacter species are non-motile and oxidase-negative, and occur in pairs under magnification.
They are important soil organisms, where they contribute to the mineralization of, for example, aromatic compounds. Acinetobacter are a key source of infection in debilitated patients in the hospital, in particular the species Acinetobacter baumannii.[1]
Etymology
Acinetobacter is a compound word from scientific Greek, meaning 'nonmotile rod'. The first element acineto- is an unusual transliteration of the Greek ακινητο-; the usual romanization in English is akineto-, as in akinetic.
Description
Species of the genus Acinetobacter are strictly aerobic nonfermentative Gram-negative bacilli. They show preponderantly a coccobacillary morphology on nonselective agar. Rods predominate in fluid media, especially during early growth.
The morphology of Acinetobacter sp. can be quite variable in Gram-stained human clinical specimens, and cannot be used to differentiate Acinetobacter from other common causes of infection.
Most strains of Acinetobacter, except some of the A. lwoffii strain, grow well on MacConkey agar (without salt). Although officially classified as nonlactose-fermenting, they are often partially lactose-fermenting when grown on MacConkey agar. They are oxidase-negative, nonmotile, and usually nitrate negative.
Taxonomy
The genus Acinetobacter comprises 17 validly named and 14 unnamed (genomic) species.[2] Some unrelated (genomic) species have common designations, while some other species seem to be congruent but have different names. The knowledge of the biology or ecology of acinetobacters at species level is limited. This is because identification of acinetobacters at species level is difficult. A phenotypic species identification system has been described, and a variety of genotypic methods has been explored and applied to investigate the diversity or phylogeny in the genus. These methods include high-resolution fingerprinting with AFLP, PCR-RFLP with digestion of PCR amplified sequences, and analysis of various DNA sequences. Of these, AFLP analysis and amplified 16SrRNA ribosomal DNA restriction analysis have been validated with large numbers of strains of all described species. Nucleotide sequence-based methods are expected to be the standard for identification in the near future.[2]
However, because routine identification in the clinical microbiology laboratory is not (yet) possible, they are divided and grouped into three main complexes:
- Acinetobacter calcoaceticus-baumanii complex: glucose-oxidising nonhemolytic, (A.baumannii can be identified by OXA-51 typing)
- Acinetobacter lwoffii: glucose-negative nonhemolytic
- Acinetobacter haemolyticus: hemolytic
Identification
Different species of bacteria in this genus can be identified using Fluorescence-Lactose-Denitrification medium (FLN) to find the amount of acid produced by metabolism of glucose.
The other reliable identification test at genus level is chromosomal DNA transformation assay (CTA): In this aasy, a naturally competent tryptophan auxotrophic mutant of Acinetobacter baylyi (BD4 trpE27) is transformed with the total DNA of a putative Acinetobacter isolate and the transformation mixture plated on a brain heart infusion agar. The growth is then harvested after incubation for 24 h at 30oC, plating on an Acinetobacter minimal agar (AMM), and incubating at 30oC for 108 h. Growth on the minimal agar medium indicates a positive transformation assay and confirmes the isolate as a member of the genus Acinetobacter. E. coli HB101 and A. calcoaceticus MTCC1921T can be used as the negative and positive controls, respectively.[3]
Natural habitat
Acinetobacter spp are widely distributed in nature. They are able to survive on various surfaces (both moist and dry) in the hospital environment, thereby being a major source of infection in debilitated patients. Occasional strains are isolated from foodstuffs, and some are able to survive on various medical equipment and even on healthy human skin. On Discovery Channel's MythBusters, hundreds of Acinetobacter colonies were discovered on an everyday kitchen sponge.
In drinking water, Acinetobacter have been shown to aggregate bacteria that otherwise do not form aggregates.
Clinical significance
In general, Acinetobacter species are considered nonpathogenic to healthy individuals. However, several species persist in hospital environments and cause severe, life-threatening infections in compromised patients. The spectrum of antibiotic resistances of these organisms together with their survival capabilities make them a threat to hospitals, as documented by recurring outbreaks both in highly developed countries and elsewhere. An important factor for their pathogenic potential is, it is presumed, an efficient means of horizontal gene transfer.
Most infections occur in immunocompromised individuals, and the strain A. baumannii is the second-most-commonly-isolated nonfermenting bacteria in human specimens.
Acinetobacter is frequently isolated in nosocomial infections and is especially prevalent in intensive care units, where both sporadic cases as well as epidemic and endemic occurrence is common. A. baumannii is a frequent cause of nosocomial pneumonia, especially of late-onset ventilator associated pneumonia. It can cause various other infections including skin and wound infections, bacteremia, and meningitis, but A. lwoffi is mostly responsible for the latter. A. baumannii can survive on the human skin or dry surfaces for weeks.
Biofilm formation is an important feature of most clinical isolates of Acinetobacter spp. Biofilms are assemblages of surface microbial cells that are enclosed in an extracellular polymeric matrix. It is clear from the epidemiologic evidence that Acinetobacter biofilms play a role in infectious diseases such as cystic fibrosis and periodontitis, and in bloodstream and UTI because of their ability to indwell medical devices. Antibiotic resistance markers are often plasmid-borne, and plasmids present in Acinetobacter strains can be transferred to other pathogenic bacteria. The ability of Acinetobacter species to adhere to the surfaces, form biofilms, and display antibiotic resistance and gene transfer means that there is an urgent need to study the factors responsible for their spread.[4]
Since the start of the Iraq War, over 700 U.S. soldiers have been infected or colonized by A. baumannii. Four civilians undergoing treatment for serious illnesses at Walter Reed Army Medical Center in Washington, D.C. contracted A. baumannii infections and died. At Landstuhl Regional Medical Center, a U.S. military hospital in Germany, another civilian under treatment, a 63-year-old German woman, contracted the same strain of A. baumannii infecting troops in the facility and also died. These infections appear to have been hospital-acquired. Based on genotyping of A. baumannii cultured from patients prior to the start of the Iraq War, one can presume that it is likely the soldiers contracted the infections while hospitalized for treatment in Europe.
Treatment
Acinetobacter species are innately resistant to many classes of antibiotics, including penicillin, chloramphenicol, and often aminoglycosides. Resistance to fluoroquinolones has been reported during therapy, which has also resulted in increased resistance to other drug classes mediated through active drug efflux. A dramatic increase in antibiotic resistance in Acinetobacter strains has been reported by the CDC and the carbapenems are recognised as the gold-standard and treatment of last resort.[5] Acinetobacter species are unusual in that they are sensitive to sulbactam; sulbactam is most commonly used to inhibit bacterial beta-lactamase, but this is an example of the antibacterial property of sulbactam itself.[6]
In November, 2004, the CDC reported an increasing number of A. baumannii bloodstream infections in patients at military medical facilities in which service members injured in the Iraq/Kuwait region during Operation Iraqi Freedom (OIF) and in Afghanistan during Operation Enduring Freedom (OEF) were treated.[7] Most of these were multidrug-resistant. Among one set of isolates from Walter Reed Army Medical Center, 13 (35%) were susceptible to imipenem only, and two (4%) were resistant to all drugs tested. One antimicrobial agent, colistin (polymyxin E), has been used to treat infections with multidrug-resistant A. baumannii; however, antimicrobial susceptibility testing for colistin was not performed on isolates described in this report. Because A. baumannii can survive on dry surfaces for up to 20 days, they pose a high risk of spread and contamination in hospitals, potentially putting immune-compromised and other patients at risk for drug-resistant infections that are often fatal and, in general, expensive to treat.
Reports suggest that this bacteria is susceptible to phage therapy.[8] A phage directed against Acinetobacter showed a remarkable lytic activity both in vitro and in vivo: as few as 100 pfu of phage protected mice against Acinetobacter.[9]
Biotechnology
Many of the characteristics of Acinetobacter ecology, taxonomy, physiology, and genetics point to the possibility of exploiting its unique features for future applications. Acinetobacter strains are often ubiquitous, exhibit metabolic versatility, and are robust. And some provide convenient systems for modern molecular genetic manipulation and subsequent product engineering. These characteristics are being exploited in various biotechnological applications including biodegradation and bioremediation, novel lipid and peptide production, enzyme engineering, biosurfactant, and biopolymer production and engineering of novel derivatives of these products. It is anticipated that progress in these fields will broaden the range of applications of Acinetobacter for modern biotechnology.[10]
References
- ^ Gerischer U (editor). (2008). Acinetobacter Molecular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-20-2.
{{cite book}}:|author=has generic name (help) - ^ a b Dijkshoorn L (2008). "The Diversity of the Genus Acinetobacter". Acinetobacter Molecular Biology (Gerischer U, ed.). Caister Academic Press. ISBN 978-1-904455-20-2.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Rokhbakhsh-Zamin F., D.P. Sachdev, N. Kazemi-Pour, A. Engineer, S.S. Zinjarde, P.K. Dhakephalkar and B.A. Chopade.(2011). Characterization of plant growth promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. J Microbiol Biotechnol. 21(6): 556-566.
- ^ Antunes LC, Imperi F, Carattoli A, Visca P. Deciphering the Multifactorial Nature of Acinetobacter baumannii Pathogenicity. PLoS One. 2011;6(8):e22674. Epub 2011 Aug 1.
- ^ Rahal J (2006). "Novel antibiotic combinations against infections with almost completely resistant Pseudomonas aeruginosa and Acinetobacter species". Clin Infect Dis. 43 Suppl 2: S95–9. doi:10.1086/504486. PMID 16894522.
- ^ Wood GC, Hanes SD, Croce MA, Fabian TC, Bougher BA. (2002). "Comparison of ampicillin-sulbactam and imipenem-cilastin for the treatment of Acinetobacter ventilator-associated pneumonia". Clin Infect Dis. 34 (11): 1425–30. doi:10.1086/340055. PMID 12015687.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Centers for Disease Control and Prevention (CDC) (2004). "Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004". MMWR Morb Mortal Wkly Rep. 53 (45): 1063–6. PMID 15549020.
- ^ Matsuzaki S, Rashel M, Uchiyama J; et al. (2005). "Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases". J. Infect. Chemother. 11 (5): 211–9. doi:10.1007/s10156-005-0408-9. PMID 16258815.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Brussow H (2007). "Phage Therapy: The Western Perspective". Bacteriophage: Genetics and Molecular Biology (Mc Grath S and van Sinderen D, ed.). Caister Academic Press. ISBN 978-1-904455-14-1.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Gutnick D (2008). "Potential Application of Acinetobacter in Biotechnology". Acinetobacter Molecular Biology (Gerischer U, ed.). Caister Academic Press. ISBN 978-1-904455-20-2.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help)
External links
- Alliance for the Prudent Use of Antibiotics
- Acinetobacter sp. ADP1 Genome Page
- Transclusion error: {{En}} is only for use in File namespace. Use {{lang-en}} or {{in lang|en}} instead. CycSim: metabolic model of Acinetobacter baylyi adp1
- Transclusion error: {{En}} is only for use in File namespace. Use {{lang-en}} or {{in lang|en}} instead. AcinetoCyc: Pathway Genome database for Acinetobacter baylyi adp1
