Glycineamide ribonucleotide
Appearance
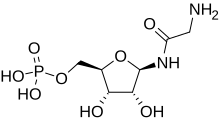
| |
| Names | |
|---|---|
| IUPAC name
(1R)-1,4-Anhydro-1-glycinamido-D-ribitol 5-(dihydrogen phosphate)
| |
| Systematic IUPAC name
[(2R,3S,4R,5R)-5-(2-Aminoacetamido)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen phosphate | |
| Other names
Glycineamide ribotide,
GAR | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
| KEGG | |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C7H15N2O8P | |
| Molar mass | 286.177 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Glycineamide ribonucleotide (or GAR) is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA.[1][2][3] The vitamins thiamine[4] and cobalamin[5] also contain fragments derived from GAR.[6]
GAR is the product of the enzyme phosphoribosylamine—glycine ligase acting on phosphoribosylamine (PRA) to combine it with glycine in a process driven by ATP. The reaction, EC 6.3.4.13 forms an amide bond:[7]
- PRA + glycine + ATP → GAR + ADP + Pi
The biosynthesis pathway next adds a formyl group from 10-formyltetrahydrofolate to GAR, catalysed by phosphoribosylglycinamide formyltransferase in reaction EC 2.1.2.2 and producing formylglycinamide ribotide (FGAR):[7]
- GAR + 10-formyltetrahydrofolate → FGAR + tetrahydrofolate
See also
[edit]References
[edit]- ^ R. Caspi (2009-01-13). "Pathway: 5-aminoimidazole ribonucleotide biosynthesis I". MetaCyc Metabolic Pathway Database. Retrieved 2022-02-02.
- ^ Zhang, Y.; Morar, M.; Ealick, S.E. (2008). "Structural biology of the purine biosynthetic pathway". Cellular and Molecular Life Sciences. 65: 3699–3724. doi:10.1007/s00018-008-8295-8. PMC 2596281. PMID 18712276.
- ^ Gupta, Rani; Gupta, Namita (2021). "Nucleotide Biosynthesis and Regulation". Fundamentals of Bacterial Physiology and Metabolism. pp. 525–554. doi:10.1007/978-981-16-0723-3_19. ISBN 978-981-16-0722-6. S2CID 234897784.
- ^ Chatterjee, Abhishek; Hazra, Amrita B.; Abdelwahed, Sameh; Hilmey, David G.; Begley, Tadhg P. (2010). "A "Radical Dance" in Thiamin Biosynthesis: Mechanistic Analysis of the Bacterial Hydroxymethylpyrimidine Phosphate Synthase". Angewandte Chemie International Edition. 49 (46): 8653–8656. doi:10.1002/anie.201003419. PMC 3147014. PMID 20886485.
- ^ R. Caspi (2019-09-23). "Pathway: 5-hydroxybenzimidazole biosynthesis (anaerobic)". MetaCyc Metabolic Pathway Database. Retrieved 2022-02-10.
- ^ Mehta, Angad P.; Abdelwahed, Sameh H.; Fenwick, Michael K.; Hazra, Amrita B.; Taga, Michiko E.; Zhang, Yang; Ealick, Steven E.; Begley, Tadhg P. (2015). "Anaerobic 5-Hydroxybenzimidazole Formation from Aminoimidazole Ribotide: An Unanticipated Intersection of Thiamin and Vitamin B12 Biosynthesis". Journal of the American Chemical Society. 137 (33): 10444–10447. doi:10.1021/jacs.5b03576. PMC 4753784. PMID 26237670.
- ^ a b Welin, Martin; Grossmann, Jörg Günter; Flodin, Susanne; Nyman, Tomas; Stenmark, Pål; Trésaugues, Lionel; Kotenyova, Tetyana; Johansson, Ida; Nordlund, Pär; Lehtiö, Lari (2010). "Structural studies of tri-functional human GART". Nucleic Acids Research. 38 (20): 7308–7319. doi:10.1093/nar/gkq595. PMC 2978367. PMID 20631005.

