BioSLAX
 Various bio apps running on BioSLAX | |
| Developer | National University of Singapore BioInformatics Center (Resource) Mark De Silva Lim Kuan Siong Tan Tin Wee |
|---|---|
| OS family | Linux |
| Working state | Current |
| Source model | Open source |
| Latest release | v 7.5 / February 5, 2009 |
| Kernel type | Monolithic kernel |
| License | Various |
| Official website | www.bioslax.com |
BioSLAX is a Live CD/Live DVD/Live USB comprising a suite of more than 300 bioinformatics tools and application suites. It has been released by the Bioinformatics Resource Unit of the Life Sciences Institute (LSI), National University of Singapore (NUS) and is bootable from any PC that allows a CD/DVD or USB boot option and runs the compressed Slackware flavour of the Linux Operating System (OS), also known as Slax. Slax was created by Tomáš Matějíček in the Czech Republic using the Linux Live Scripts which he also developed. The BioSLAX derivative was created by Mark De Silva, Lim Kuan Siong and Tan Tin Wee.
BioSLAX was first released to the NUS Life Science Curriculum in April 2006.
History
In January 2003, APBioNet received a research grant from the Pan Asia Networking (PAN) Programme of IDRC (Canada) to build an APBioBox of commonly used bioinformatics applications and packages with grid-computing software as part of its effort to build an APBioGrid. The platform chosen was the then ubiquitous Redhat Linux. In March of that same year, APBioNet launched an industry partnership scheme (AIPS) and partnered with Sun Microsystems to build BioBox for the Solaris platform. Six months later, beta versions of APBioBox and Sun's biobox, now called Bio-Cluster Grid were released for beta testing among selected parties. The packages included Globus Grid Toolkit Version 2.0 and Sun Grid Engine respectively.[1]
On 4 December 2003, the biobox software packages now named APBioBox (Redhat Linux) and BioCluster Grid (Sun Solaris) were field-tested at a Bioinformatics Workshop was conducted at the Advanced Science and Technology Institute (ASTI), Department of Science and Technology (DOST), Philippines on the occasion of the 70th Anniversary of the National Research Council of the Philippines (NRCP). Ten pentium machines and a couple of Sun servers were successfully inducted into the APBioGrid. This Workshop and the software tested were sponsored by Sun Microsystems and partially funded by IDRC.
In July 2004, Dr Derek Kiong introduced Knoppix as a stable, powerful and small-footprint Unix (Debian-based) platform to A/Prof Tan Tin Wee in a workshop organised by the Institute of Systems Science (ISS), NUS. By September 2004, through Mr Ong Guan Sin, we were able to create a Knoppix remaster template by building software in APBioBox plus useful applications into a prototype, APBioKnoppix, as a project for the practical course of LSM2104 module of the Dept of Biochemistry, NUS.[2] It was subsequently upgraded based on Knoppix 4.02 and released as APBioKnoppix2.[3] While APBioKnoppix was widely used, it was found that it was not easily expandable. All applications had to be in place prior to remastering and this made the distribution highly inflexible.
In June 2005, Mr. Mark De Silva of the Bioinformatics Resource Unit of the Life Sciences Institute (LSI), suggested using Slax as a base for a new bio-based live CD due to its modular system, which effectively allowed for the same base system to be used and various tools or changes to be included on top of the base easily by adding single modules with all the application files or changes. This eliminated the need to remaster the entire system every time new software or changes emerged, which was the case for Knoppix.
By April 2006, the first version of BioSLAX was released with several editions:
- Standard User Edition (530 MBytes)
- Developer Edition (700 MBytes)
- Sever Edition (470 MBytes)
BioSLAX was subsequently used in the bioinformatics teaching module within NUS under the Life Science Curriculum as well as in several events that were organized under the umbrella of the Asia Pacific Bioinformatics Network (APBioNet). APBioNet is a regional affiliate of the International Society for Computational Biology (ISCB). Customized versions were built to cater for both NUS and APBioNet.
In August 2007, in collaboration with the APBioNet, a customized BioSLAX was used to set up the Bioinformatics Resource Node of Vietnam at Bio-IBT, the Bioinformatics Resource Server of the Institute of Biotechnology, Vietnam Academy of Science and Technology, Hanoi, Viet Nam. The Bio-IBT node offered :
- BioMirrors repository of biological databases
- NCBI BLAST mirrored resource
- Web access to EBI EMBOSS applications
- Web access to CLUSTALW multiple sequence alignment
- Web access to the T-Coffee multiple sequence alignment
- Web access to the PHYLIP Phylogenetic Inference Package
- Web access to the Sequence Manipulation Suite, SMS2
Users with SSH access to the server also had access to many more command line based bio/life science applications.
The entire project was done in collaboration with the 1st UNESCO-IUBMB-FAOBMB-APBioNet Bioinformatics Workshop in Vietnam, held 20th to 31 August 2007, a satellite event of the 6th International Conference on Bioinformatics (InCoB) 2007 at HongKong, Hanoi and Nansha.
Some versions of BioSLAX deployed in international institutions under APBioNet were fitted with a small tool which allowed them to map their IPs to a dynamically created apbionet.org domain name, hence giving each machine a fully qualified domain name (FQDN) and presence on the Internet.[4]
Modularity
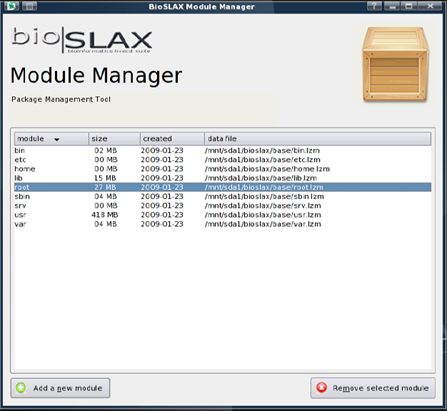
Because Slax worked by overlaying "application modules" on top of the base Linux OS, it made the entire distribution modular. The additional functionality of deploying these modules even while the system was already running, made using Slax even more appealing. The inclusion of the GUI based "BioSLAX Module Manager", made this process of dynamically adding and removing modules even easier.
Users were able to test updates to software or new versions and "rollback" to previous versions if they want. This was especially effective if SLAX/BioSLAX was installed to a writable medium such as a USB drive.
Versions
To date, there have been two versions of BioSLAX - BioSLAX 5.x based on Slax 5 and BioSLAX 7.x based on Slax 6. While BioSLAX 5.x followed the version numbers of Slax 5, BioSLAX 7 adopted a new version numbering which is one higher than the Slax version on which it is based. Latest versions can be downloaded from the BioSLAX website.[5]
BioSLAX 5.x
BioSLAX 5.x was largely based on the 5.1.8 version of Slax, running earlier versions of the 2.6 Linux kernel and KDE 3.4, with unionfs.
BioSLAX 5.x Editions
Standard User Edition
This edition runs the KDE X Window GUI and comes with all the tools and application suites, but does not include any compiler tools nor the Linux kernel source code and headers. This is mainly suited for users who only need to use the tools and applications suites. It has a very small size, making it easy to download and particularly convenient for regions where internet bandwidth is an issue.
Developer Edition
This edition runs the KDE X Window GUI and comes with all the tools and application suites and also includes a full set of development and compiler tools and also including the Linux kernel source code and headers. This is edition is more for the power user, who, in addition to using the various tools and applications, might want to also compile new applications or create new application modules for BioSLAX.
Server Edition
This edition does not include any X Window GUI, compilation tools, Linux kernel source or kernel headers. It is primarily meant to be used as a remote server, where users have to either SSH in to use the command line applications or connect to the server via the web to access the available web-based portals to popular bio applications.
NUS LSM Edition
This edition is the Developer Edition, customized for use by the NUS Life Science Curriculum for the teaching of bioinformatics.
Taverna Edition
This edition is the Developer Edition which includes TaveRNA. The TaveRNA Project aims to provide a language and software tools to facilitate easy use of workflow and distributed compute technology.
BioSLAX 7.x
BioSLAX 7.x is based on Slax 6 and features the later releases of the 2.6 Linux kernel, KDE 3.5 and using aufs and lzma compression. The biggest change is the use of this version as either client or server. The distribution was also moved from CD to DVD, allowing for more applications to be introduced, which were previously left out of version 5.x due to space considerations. The ability to boot from a FAT or EXT formatted USB drive was also introduced in Slax 6, hence BioSLAX 7.x versions also had this feature, effectively enabling persistent file handling which are unavailable on the CD/DVD as they are not (re-)writable.
BioSLAX 8
Versions of BioSLAX after 7.x have been delayed due to the base distribution's (Slax) developer, Tomáš Matějíček, refusing to move forward with a new version because of family commitments. However his primary reason for not moving forward was that he was waiting for Squash FS and LZMA to be integrated into the Linux kernel by default, instead of users needing to apply separate patches. As of kernel 2.6.38, the integration was finally done and this has prompted Tomáš Matějíček to look at a new version of Slax, which will therefore result in a new version of BioSLAX in the coming months. One can follow his thoughts on the new version of Slax on his blog[permanent dead link].
Features
Standard Tools
BioSLAX features the Linux Slackware 12.1 operating system with updated drivers for various network adapters including support for a large variety of wireless cards. It also has many useful basic tools and applications such as:
- PERL (including BioPerl modules)
- PHP
- Apache 2
- MySQL
- OpenOffice.org
- KPDF Reader
- Mozilla Firefox
- Mozilla Thunderbird
- gFTP
- ProFTPd
- Open SSH
- Kopete Instant Messenger
- VNC Viewer
- Remote Desktop Services
BioInformatics Tools
The bioinformatics tools and applications are subdivided into three main categories.
Console Apps
- BLAST
- BlastCL3
- BioGrep
- ClustalW
- EMBOSS
- Genesplicer
- GlimmerHMM
- HMMER
- Modeller
- PamL
- Phylip
- Primer3
- R programming language & Bioconductor
- T-Coffee
Desktop Apps
- ACT
- Artemis
- ClustalX (GUI Based ClustalW)
- JAligner
- Jalview
- jEMBOSS (Java EMBOSS Suite)
- Jmol
- NJPlot
- Pymol
- ReadSEQ
- TreeView
- Weka (machine learning)
Web Apps
- Web BLAST
- Web ClustalW
- Web Phylip
- Web T-Coffee
- wEMBOSS (Web based EMBOSS suite)
- Sequence Manipulation Suite (SMS)
Installing to Hard Disk
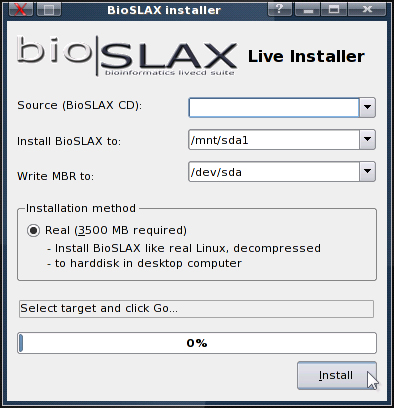
One of the more intriguing features of Slax-based distributions is how easy it is to convert the live OS into a full-fledged Linux system installed on the hard drive of any PC, which will take up roughly 3.5GBytes of space.
A tool, written with the KDE Kommander toolkit called the "BioSLAX Installer" is provided for users to easily convert their live OS to a full Linux installation. By using modules to customize the distribution and then using the installer, users can do rapid deployment of fully installed customized clients.
Future plans
BioSLAX Updates
BioSLAX will be updated as newer Slackware (or Slax) versions are released. The tools and applications suites will also be monitored for significant changes and upgraded as necessary. Some tools may be removed to make way for other tools which can do the same thing but with added functionality and better efficiency. More web-based portals are being looked at, for example, portals to ReadSeq, Primer3 and Genesplicer are in the pipeline.
Grid Deployment
The developers were also looking at integrating various Grid computing platforms with BioSLAX. Because BioSLAX can be booted up immediately from any CD/DVD/USB, it can be used as a rapidly deployable Grid-enabled Operating System. One such Grid platform was the Univa Grid platform. Using the Univa Grid MP agent, it was shown during GridAsia 2009 in a talk given by Tan Tin Wee, that the agent, once modularized on BioSLAX, can be used to Grid enable machines from any location as slave-nodes to a master-node located elsewhere, effectively creating a "global-wide grid".
BioSLAX on the CLOUD
In a proof-of-concept endeavour, the developers successfully deployed BioSLAX as instances on a pool of resources using both VMWare's ESXi and Citrix Xen's Hypervisors. Their aim was to effectively create a "BioSLAX CLOUD" where students and staff may instantiate any number BioSLAX servers dynamically for research and education (conduct bioinformatics practical labs by having students connect to the servers via suitable X Window clients such as X-Win32, VNC, Exceed and NoMachine NX) or deployed in such a manner which when used in conjunction with the UD Grid mpagent may be used to form a cluster for processing large jobs.
The proof-of-concept was highly successful in being deployed for research and education for the Life Science Curriculum at NUS and in 2011, a number of the BioSLAX cloud instances, both on VMWare's vSphere and Citrix Xen servers, were used in the APBioNet project, BioDB100. The backend controls and automation were created and implemented using the various APIs for vSphere and Xen by Mr. Mark De Silva.
Developers were also in talks with Amazon from 2009 to 2010 to deploy similar BioSLAX cloud images on Amazon's EC2, hoping to push some of their research and education machines over to Amazon, cutting costs on hardware. Discussions, however, fell through when it was clear that Amazon was not going to support full hardware virtualization which was required in order to run BioSLAX images on the cloud. Supporting only para-virtualizaion, in fact, is the stand of most commercial cloud providers using Citrix Xen hypervisors. Until the mind-set of these entities change, only private clouds running Citrix Xen hypervisors configured for full hardware virtualization or VMWare vSphere clouds will be the only clouds capable of running BioSLAX.