CCDC138
Coiled-coil domain-containing protein 138, also known as CCDC138, is a human protein encoded by the CCDC138 gene. The exact function of CCDC138 is unknown.
Gene
The CCDC138 gene can be found at the positive strand of chromosome 2.[5]
Locus
The CCDC138 gene is located at the long(q) arm of chromosome 2 at locus 12.13,[6] or in short 2q12.3. It can be found at location 108,786,752-108,876,591.[7] The DNA sequence is 89,840bp long.

Common aliases
CCDC138 is the only established common alias.
Homology and evolution
Paralogs
No paralogs of CCDC138 have been identified.
Orthologs
CCDC138 is conserved in various organisms as shown in the table below.
| Scientific name | Common name | Date of divergence from human lineage[8] | Sequence length | Sequence identity to human RNA/protein | Sequence similarity to human RNA/protein |
|---|---|---|---|---|---|
| Mus musculus | House mouse | 92.3 MYA | 2466 bp | 77% | 65.7% |
| Columbia livia | Rock dove | 296 MYA | 1863 bp | 61% | 59.4% |
| Xenopus laevis | African clawed frog | 371.2 MYA | 2634 bp | 52% | 40.1% |
| Anolis carolinensis | Red-throated anole | 296 MYA | 9588 bp | 78% | 46.3% |
| Latimeria chalumnae | West Indian Ocean coelacanth | 414.9 MYA | 1838 bp | 71% | 38.1% |
| Strongylocentrotus purpuratus | Purple sea urchin | 742.9 MYA | 2047 bp | 59% | 14.5% |
| Ciona intestinalis | Vase tunicate | 722.5 MYA | 2420 bp | 56% | 23.8% |
| Aplysia californica | California sea slug | 782.7 MYA | 2103 bp | 49% | 17.2% |
| Hydra vulgaris | Fresh-water polyp | 855.3 MYA | 1482 bp | 46% | 13.7% |
| Chrysemys picta bellii | Western painted turtle | 296 MYA | 700 bp | 36% | 15.9% |
| Alligator mississippiensis | American alligator | 296 MYA | 2089 bp | 76% | 38.4% |
| Melopsittacus undulatus | Budgerigar | 296 MYA | 1764 bp | 73% | 40.3% |
| Taeniopygia guttata | Zebra finch | 296 MYA | 1980 bp | 75% | 44.2% |
| Lepisosteus oculatus | Spotted gar | 400.1 MYA | 1269 bp | 65% | 30.9% |
| Saccoglossus kowalevskii | Acorn worm | 661.2 MYA | 2515 bp | 42% | 24.1% |
| Branchiostoma floridae | Lancelet | 713.2 YA | 1758 bp | 55% | 27.0% |
| Maylandia zebra | Zebra mbuna fish | 400.1 MYA | 4815 bp | 58% | 21.4% |
| Trichoplax adhaerens | Trichoplax | 800 MYA | 1605 bp | 56% | 10.3% |
| Pelodiscus sinensis | Chinese softshell turtle | 296 MYA | 2895 bp | 77% | 33.9% |
| Falco cherrug | Saker falcon | 296 MYA | 1866 bp | 73% | 48.9% |
Distant homologs
The most distant homolog detected or predicted is Trichoplax adhaerans. It has a conserved CCDC138 gene and has evolved 800 MYA before the human lineage.
Homologous domains
Among the orthologs stated above, there are various homologous regions that are conserved shown in figures below.
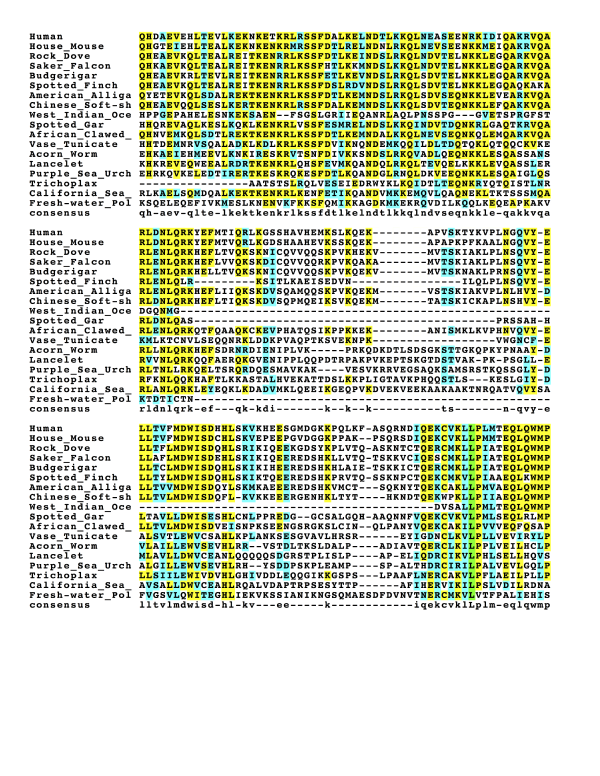


Green colors shows completely conserved residues, yellow color shows identical residues, cyan color shows similar residues, white color shows different residues.
Phylogeny
The observed phylogeny of the CCDC138 gene of the above mentioned orthologs recapitulates the evolutionary history.[9]

The figure above shows the evolutionary relationship of CCDC138 in the orthologs.
Protein
The CCDC138 protein is predated to have a molecular weight of 76.2Kda[10] and an isoelectric point of 8.614.[11] Compositional analysis shows that there is a low usage of the AGP grouping in CCDC138, and there are no positive, negative or mixed charge clusters. The protein has no ER retention motif in the C-terminus and no RNA binding motif.[12] It has also been predicted to be a soluble nuclear protein with a leucine zipper pattern (PS00029) at position 205 onwards with a sequence LQKRERFLLEREQLLFRHENAL.[12]
Primary sequence and variants/isoforms
There are two isoforms of the CCDC138 protein. The primary isoform has 665 amino acids[13] while the secondary isoform has 577 amino acids,[13] and is missing 88 amino acids at the C-terminus.

Figure shows the pairwise sequence alignment comparing the primary isoform (Isoform 1) to the secondary isoform (Isoform 2).
Domain and motifs
A domain of unknown function (DUF2317) on the protein at location 212 – 315 has been characterized in bacteria. TMHMM[14] and TMAP[15] suggests that there are no predicted transmembrane domain. SOSUI[16] further predicts that CCDC138 is a soluble protein with no transmembrane domain.
Post-translational modifications
According to SUMOplot Analysis Program,[17] there are 7 predicted sumoylation at lysine residues K7, K207, K336, K374, K383, K521, and K591. NetPhos[18] predicts that there are 44 phosphorylations sites, including 29 serine residues, 10 threonine residues, and 5 tyrosine residues. There are no further post-translational modifications as predicted by NetNGlyc,[19] NetOGlyc,[20] SignalP,[21] Sulfinator,[22] and Myristoylator.[23]
Secondary structure
The CCDC138 protein contains multiple alpha helixes, beta sheets and coiled-coils as predicted by PELE, CHOFAS, and GOR4.

Yellow shows coiled-coil, blue shows alpha helix, and red shows beta sheet. The majority of the sequence are coiled-coils and alpha helixes.
3° and 4° structures
There are no predicted 3° and 4° Structures for the CCDC138 protein. However, there is a similar structure that has a 29% identity.[24] The predicted structure is Chain A, crystal structure analysis of Clpb, a protein that encodes an ATP-dependent protease and chaperone. This protein has an aligned-length of 144 amino acids, and the alignment is located at the domain of unknown function of CCDC138.
Expression
The gene is expressed at low levels in almost all human tissues, but higher levels have been seen in certain cancer tissues. CCDC138 is a soluble protein that is pre diced to localise in the nucleus of a cell.
Promoter
The promoter region of CCDC138 is shown as figure below.

Expression
Microarray-assessed tissue expression patterns through GEO profiles show that CCDC138 is expressed in moderate levels in various tissues including peripheral blood lymphocyte, fetal thymus, thymus, testis, ovary, feral brain, colon, mammary gland, and bone marrow.[25]

Transcript variants
There are two most significant alternative transcript variants for CCDC138 mRNA. The first variant as shown in the figure below has been found in lung, blood, and human embryonic stem cells.[26] The second variant has been found in adenocarcinoma, prostate, lung, and primary lung epithelial cells.[27]

First transcript shows the complete mRNA transcript. Second transcript is the first variant, while the thirst transcript is the second variant.[28]
Function and biochemistry
The exact function of CCDC138 is yet to be known.
Interacting proteins
The CCDC138 protein has been found to interact with ubiquitin C,[29] a protein involved in ubiquination and eventually protein degradation.
Transcription factors that might bind to regulatory sequence
The table below shows some transcription factors that have been predicted by Genomatix that binds to the regulatory sequence of the CCDC138 gene.[30]
| Detailed family information | Detailed matrix information | Tissue |
|---|---|---|
| GC-Box factors SP1/GC | Stimulating protein 1, ubiquitous zinc finger transcription factor | Ubiquitous |
| Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma, DR1 sites | Adipose Tissue, Connective Tissue, Digestive System, Liver |
| MYT1 C2HC zinc finger protein | MyT1 zinc finger transcription factor involved in primary neurogenesis | Central Nervous System, Nervous System, Neuroglia, Neurons |
| NGFI-B response elements, nur subfamily of nuclear receptors | Monomers of the nur subfamily of nuclear receptors (nur77, nurr1, nor-1) | Brain, Central Nervous System, Endocrine System, Immune System, Leydig Cells, Nervous System, Neurons, Testis, Thymus Gland, Urogenital System |
| Krueppel-like transcription factors | Core promoter-binding protein (CPBP) with 3 Krueppel-type zinc fingers (KLF6, ZF9) | Blood cells, bone marrow cells, digestive system, embryonic structures, Erythrocytes, Hematopoietic System, liver |
| Grainyhead-like transcription factors | Grainyhead-like 3 (sister-of-mammalian grainyhead - SOM) | Embryonic Structures, Integumentary System |
| CTCF and BORIS gene family, transcriptional regulators with 11 highly conserved zinc finger domains | Insulator protein CTCF (CCCTC-binding factor) | Blood Cells, Embryonic Structures, Endocrine System, Erythrocytes, Germ Cells, Testis, Urogenital System |
| Core promoter motif ten elements | Human motif ten element | - |
| Abdominal-B type homeodomain transcription factors | Homeobox C13 / Hox-3gamma | Bone Marrow Cells, Bone and Bones, Central Nervous System, Connective Tissue, Embryonic Structures, Hematopoietic System, Integumentary System, Kidney, Nervous System, Neurons, Prostate, Skeleton, Spinal Cord, Urogenital System |
| E2F-myc activator/cell cycle regulator | E2F transcription factor 2 | Ubiquitous |
| PAX-3 binding sites | Pax-3 paired domain protein, expressed in embryogenesis, mutations correlate to Waardenburg Syndrome | Embryonic structures, muscle, skeletal, muscles |
| ZF5 POZ domain zinc finger | ZF5 POZ domain zinc finger, zinc finger protein 161 | - |
| Vertebrate TATA binding protein factor | Cellular and viral TATA box elements | - |
| CCAAT binding factors | Avian C-type LTR CCAAT box | Ubiquitous |
| Ccaat/Enhancer Binding Protein | CCAAT/enhancer binding protein alpha | Adipose Tissue, Bone Marrow Cells, Connective Tissue, Digestive System, Hematopoietic System, Immune System, Liver, Myeloid Cells, Phagocytes |
| Activator-, mediator- and TBP-dependent core promoter element for RNA polymerase II transcription from TATA-less promoters | X gene core promoter element 1 | - |
Clinical significance
CCDC138 has been identified as one of the many genes involved in initiating term labor in myometrium.[31]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000163006 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000038010 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Homo sapiens coiled-coil domain containing 138 (CCDC138), mRNA". Retrieved 2 Feb 2014.
- ^ "CCDC138 - GeneCards". Retrieved 2 Feb 2014.
- ^ "CCDC138 coiled-coil domain containing 138 [ Homo sapiens (human) ]". Retrieved 2 Feb 2014.
- ^ "TimeTree :: The Timescale of Life". Retrieved 5 March 2014.
- ^ "CLUSTALW". Retrieved 1 March 2014.[permanent dead link]
- ^ "SAPS". Retrieved 10 April 2014.[permanent dead link]
- ^ "pI". Retrieved 10 April 2014.[permanent dead link]
- ^ a b "PSORT WWW Server". Retrieved 18 April 2014.
- ^ a b "Coiled-coil domain-containing protein 138 - CCDC138 - Homo sapiens (Human)". Retrieved 10 February 2014.
- ^ "TMHMM". Retrieved 20 April 2014.[permanent dead link]
- ^ "TMAP". Retrieved 20 April 2014.[permanent dead link]
- ^ "Classification and Secondary Structure Prediction of Membrane Proteins". Retrieved 15 April 2014.
- ^ "SUMOplot™ Analysis Program". Retrieved 15 April 2014.
- ^ "NetPhos 2.0 Server". Retrieved 15 April 2014.
- ^ "NetNGlyc 1.0 Server". Retrieved 15 April 2014.
- ^ "NetOGlyc 4.0 Server". Retrieved 15 April 2014.
- ^ "SignalP 4.1 Server". Retrieved 15 April 2014.
- ^ "The Sulfinator". Retrieved 15 April 2014.
- ^ "Myristoylator". Retrieved 15 April 2014.
- ^ "CBLAST". Retrieved 22 April 2014.
- ^ "GDS3113 / 172084 / CCDC138". Retrieved 29 March 2014.
- ^ "AHomo sapiens coiled-coil domain containing 138 (65.7 kD) (CCDC138) alternative variant dAug10, complete mRNA". Retrieved 30 March 2014.
- ^ "Homo sapiens coiled-coil domain containing 138 (47.0 kD) (CCDC138) alternative variant fAug10, complete mRNA". Retrieved 30 March 2014.
- ^ "AceView: Gene:CCDC138, a Comprehensive Annotation of Human, Mouse and Worm Genes with mRNAs or ESTs". Retrieved 30 March 2014.
- ^ "CCDC138 protein (Homo sapiens) - STRING network view". Retrieved 22 April 2014.
- ^ "Transcription Factors". Retrieved 27 March 2014.[permanent dead link]
- ^ Weiner CP, Mason CW, Dong Y, Buhimschi IA, Swaan PW, Buhimschi CS (May 2010). "Human effector/initiator gene sets that regulate myometrial contractility during term and preterm labor". American Journal of Obstetrics and Gynecology. 202 (5): 474.e1–20. doi:10.1016/j.ajog.2010.02.034. PMC 2867841. PMID 20452493.
External links
- Human CCDC138 genome location and CCDC138 gene details page in the UCSC Genome Browser.




