CBS domain
| CBS domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
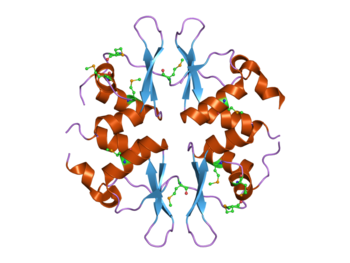 Structure of the yeast SNF4 protein that contains four CBS domains.[1] This protein is part of the AMP-activated protein kinase (AMPK) complex. | |||||||||||
| Identifiers | |||||||||||
| Symbol | CBS | ||||||||||
| Pfam | PF00571 | ||||||||||
| InterPro | IPR000644 | ||||||||||
| SMART | CBS | ||||||||||
| PROSITE | PS51371 | ||||||||||
| SCOP2 | 1zfj / SCOPe / SUPFAM | ||||||||||
| CDD | cd02205 | ||||||||||
| |||||||||||
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in.[2] CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase,[3] voltage gated chloride channels[4][5][6][7][8] and AMP-activated protein kinase (AMPK).[9][10] CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.[11]
Structure
The CBS domain is composed of a beta-alpha-beta-beta-alpha secondary structure pattern that is folded into a globular tertiary structure that contains a three-stranded antiparallel β-sheet with two α-helices on one side. CBS domains are always found in pairs in protein sequences and each pair of these domains tightly associate in a pseudo dimeric arrangement through their β-sheets forming a so-called CBS-pair or Bateman domain.[12][13] These CBS domain pairs can associate in a head-to-head (i.e. PDB codes, 3KPC, 1PVM, 2OOX) or a head-to-tail (i.e. PDB codes 1O50, 1PBJ) manner forming a disk-like compact structure. By doing so, they form clefts that constitute the canonical ligand binding regions.[14][15][16][17][18] In principle, the number of canonical binding sites matches the number of CBS domains within the molecule and are traditionally numbered according to the CBS domain that contains each of the conserved aspartate residues that potentially interact with the ribose of the nucleotides.[19] However, not all of these cavities might necessarily bind nucleotides or be functional. Recently, a non-canonical site for AMP has also been described in protein MJ1225 from M. jannaschii, though its functional role is still unknown.[20]

Ligand binding
It has been shown that CBS domains bind to adenosyl groups in molecules such as AMP and ATP,[11] or s-adenosylmethionine,[21] but they may also bind metallic ions such as Mg2+.[22][23] Upon binding these different ligands the CBS domains regulate the activity of associated enzymatic domains.[24] The molecular mechanisms underlying this regulation are just starting to be elucidated.[16][17][21][22][25] At the moment, two different type of mechanisms have been proposed. The first one claims that the nucleotide portion of the ligand induces essentially no change in the protein structure, the electrostatic potential at the binding site being the most significant property of adenosine nucleotide binding.[17][26] This "static" response would be involved in processes in which regulation by energy charge would be advantageous.[17][26] On the contrary, the second type of mechanism (denoted as "dynamic") involves dramatic conformational changes in the protein structure upon ligand binding and has been reported for the cytosolic domain of the Mg2+ transporter MgtE from Thermus thermophilus,[22] the unknown function protein MJ0100 from M. jannaschii [21][27] and the regulatory region of Clostridium perfringens pyrophosphatase.[28]
Associated domains
CBS domains are often found in proteins that contain other domains. These domains are usually enzymatic, membrane transporters or DNA-binding domains. However, proteins that contain only CBS domains are also often found, particularly in prokaryotes. These standalone CBS domain proteins might form complexes upon binding to other proteins such as kinases to which they interact with and regulate.

Mutations leading to disease
Mutations in some human CBS domain-containing proteins leads to genetic diseases.[3] For example, mutations in the cystathionine-beta-synthase protein lead to an inherited disorder of the metabolism called homocystinuria (Template:OMIM3).[29] Mutations in the gamma subunit of the AMPK enzyme have been shown to lead to familial hypertrophic cardiomyopathy with Wolff-Parkinson-White syndrome (Template:OMIM3). Mutations in the CBS domains of the IMPDH enzyme lead to the eye condition retinitis pigmentosa (Template:OMIM3).
Humans have a number of voltage-gated chloride channel genes, and mutations in the CBS domains of several of these have been identified as the cause of genetic diseases. Mutations in CLCN1 lead to myotonia (Template:OMIM3),[30] mutations in CLCN2 can lead to idiopathic generalised epilepsy (Template:OMIM3), mutations in CLCN5 can lead to Dent's disease (Template:OMIM3), mutations in CLCN7 can lead to osteopetrosis (Template:OMIM3),[31] and mutations in CLCNKB can lead to Bartter syndrome (Template:OMIM3).
References
- ^ PDB: 2nye; Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L (January 2007). "Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding". Structure. 15 (1): 65–74. doi:10.1016/j.str.2006.11.014. PMID 17223533.
- ^ Bateman A (January 1997). "The structure of a domain common to archaebacteria and the homocystinuria disease protein". Trends Biochem. Sci. 22 (1): 12–3. doi:10.1016/S0968-0004(96)30046-7. PMID 9020585.
- ^ a b Ignoul S, Eggermont J (December 2005). "CBS domains: structure, function, and pathology in human proteins". Am. J. Physiol., Cell Physiol. 289 (6): C1369–78. doi:10.1152/ajpcell.00282.2005. PMID 16275737.
- ^ Ponting CP (March 1997). "CBS domains in CIC chloride channels implicated in myotonia and nephrolithiasis (kidney stones)". J. Mol. Med. 75 (3): 160–3. PMID 9106071.
- ^ Meyer S, Dutzler R (February 2006). "Crystal structure of the cytoplasmic domain of the chloride channel ClC-0". Structure. 14 (2): 299–307. doi:10.1016/j.str.2005.10.008. PMID 16472749.
- ^ Yusef YR, Zúñiga L, Catalán M, Niemeyer MI, Cid LP, Sepúlveda FV (April 2006). "Removal of gating in voltage-dependent ClC-2 chloride channel by point mutations affecting the pore and C-terminus CBS-2 domain". J. Physiol. (Lond.). 572 (Pt 1): 173–81. doi:10.1113/jphysiol.2005.102392. PMC 1779660. PMID 16469788.
- ^ Markovic S, Dutzler R (June 2007). "The structure of the cytoplasmic domain of the chloride channel ClC-Ka reveals a conserved interaction interface". Structure. 15 (6): 715–25. doi:10.1016/j.str.2007.04.013. PMID 17562318.
- ^ Meyer S, Savaresi S, Forster IC, Dutzler R (January 2007). "Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5". Nat. Struct. Mol. Biol. 14 (1): 60–7. doi:10.1038/nsmb1188. PMID 17195847.
- ^ Day P, Sharff A, Parra L, et al. (May 2007). "Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP". Acta Crystallogr. D. 63 (Pt 5): 587–96. doi:10.1107/S0907444907009110. PMID 17452784.
- ^ Rudolph MJ, Amodeo GA, Iram SH, et al. (January 2007). "Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding". Structure. 15 (1): 65–74. doi:10.1016/j.str.2006.11.014. PMID 17223533.
- ^ a b Kemp BE (January 2004). "Bateman domains and adenosine derivatives form a binding contract". J. Clin. Invest. 113 (2): 182–4. doi:10.1172/JCI20846. PMC 311445. PMID 14722609.
- ^ Kemp BE (January 2004). "Bateman domains and adenosine derivatives form a binding contract". J. Clin. Invest. 113 (2): 182–4. doi:10.1172/JCI20846. PMC 311445. PMID 14722609.
- ^ Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR (April 1999). "Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase". Biochemistry. 38 (15): 4691–700. doi:10.1021/bi982858v. PMID 10200156.
- ^ Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L (January 2007). "Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding". Structure. 15 (1): 65–74. doi:10.1016/j.str.2006.11.014. PMID 17223533.
- ^ Meyer S, Savaresi S, Forster IC, Dutzler R (January 2007). "Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5". Nat. Struct. Mol. Biol. 14 (1): 60–7. doi:10.1038/nsmb1188. PMID 17195847.
- ^ a b Amodeo GA, Rudolph MJ, Tong L (September 2007). "Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1". Nature. 449 (7161): 492–5. doi:10.1038/nature06127. PMID 17851534.
- ^ a b c d Townley R, Shapiro L (March 2007). "Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase". Science. 315 (5819): 1726–9. doi:10.1126/science.1137503. PMID 17289942.
- ^ Jin X, Townley R, Shapiro L (October 2007). "Structural insight into AMPK regulation: ADP comes into play". Structure. 15 (10): 1285–95. doi:10.1016/j.str.2007.07.017. PMID 17937917.
- ^ Kemp BE, Oakhill JS, Scott JW (October 2007). "AMPK structure and regulation from three angles". Structure. 15 (10): 1161–3. doi:10.1016/j.str.2007.09.006. PMID 17937905.
- ^ Gómez-García I, Oyenarte I, Martínez-Cruz LA (May 2010). "The Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii Shows Strong Conservation of Key Structural Features Seen in the Eukaryal gamma-AMPK". J Mol Biol. 399 (1): 53–70. doi:10.1016/j.jmb.2010.03.045. PMID 20382158.
- ^ a b c Lucas M, Encinar JA, Arribas EA, Oyenarte I, García IG, Kortazar D, Fernández JA, Mato JM, Martínez-Chantar ML, Martínez-Cruz LA (February 2010). "Binding of S-methyl-5'-thioadenosine and S-adenosyl-L-methionine to protein MJ0100 triggers an open-to-closed conformational change in its CBS motif pair". J. Mol. Biol. 396 (3): 800–20. doi:10.1016/j.jmb.2009.12.012. PMID 20026078.
- ^ a b c Ishitani R, Sugita Y, Dohmae N, Furuya N, Hattori M, Nureki O (October 2008). "Mg2+-sensing mechanism of Mg2+ transporter MgtE probed by molecular dynamics study". Proc. Natl. Acad. Sci. U.S.A. 105 (40): 15393–8. doi:10.1073/pnas.0802991105. PMC 2563093. PMID 18832160.
- ^ Hattori M, Nureki O (March 2008). "[Structural basis for the mechanism of Mg2 homeostasis by MgtE transporter]". Tanpakushitsu Kakusan Koso (in Japanese). 53 (3): 242–8. PMID 18326297.
- ^ Scott JW, Hawley SA, Green KA, et al. (January 2004). "CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations". J. Clin. Invest. 113 (2): 274–84. doi:10.1172/JCI19874. PMC 311435. PMID 14722619.
- ^ Tuominen H, Salminen A, Oksanen E, Jämsen J, Heikkilä O, Lehtiö L, Magretova NN, Goldman A, Baykov AA, Lahti R (May 2010). "Crystal structures of the CBS and DRTGG domains of the regulatory region of Clostridiumperfringens pyrophosphatase complexed with the inhibitor, AMP, and activator, diadenosine tetraphosphate". J. Mol. Biol. 398 (3): 400–13. doi:10.1016/j.jmb.2010.03.019. PMID 20303981.
- ^ a b Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ (September 2007). "Structural basis for AMP binding to mammalian AMP-activated protein kinase". Nature. 449 (7161): 496–500. doi:10.1038/nature06161. PMID 17851531.
- ^ Lucas M, Kortazar D, Astigarraga E, et al. (October 2008). "Purification, crystallization and preliminary X-ray diffraction analysis of the CBS-domain pair from the Methanococcus jannaschii protein MJ0100". Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64 (Pt 10): 936–41. doi:10.1107/S1744309108027930. PMC 2564890. PMID 18931440.
- ^ Tuominen H, Salminen A, Oksanen E, et al. (May 2010). "Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridium perfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate". J Mol Biol. 398 (3): 400–413. doi:10.1016/j.jmb.2010.03.019. PMID 20303981.
- ^ Shan X, Dunbrack RL, Christopher SA, Kruger WD (March 2001). "Mutations in the regulatory domain of cystathionine beta synthase can functionally suppress patient-derived mutations in cis". Hum. Mol. Genet. 10 (6): 635–43. doi:10.1093/hmg/10.6.635. PMID 11230183.
- ^ Pusch M (April 2002). "Myotonia caused by mutations in the muscle chloride channel gene CLCN1". Hum. Mutat. 19 (4): 423–34. doi:10.1002/humu.10063. PMID 11933197.
- ^ Cleiren E, Bénichou O, Van Hul E, et al. (December 2001). "Albers-Schönberg disease (autosomal dominant osteopetrosis, type II) results from mutations in the ClCN7 chloride channel gene". Hum. Mol. Genet. 10 (25): 2861–7. doi:10.1093/hmg/10.25.2861. PMID 11741829.
