Chemotaxis

Chemotaxis (from chemo- + taxis) is the movement of an organism or entity in response to a chemical stimulus.[1] Somatic cells, bacteria, and other single-cell or multicellular organisms direct their movements according to certain chemicals in their environment. This is important for bacteria to find food (e.g., glucose) by swimming toward the highest concentration of food molecules, or to flee from poisons (e.g., phenol). In multicellular organisms, chemotaxis is critical to early development (e.g., movement of sperm towards the egg during fertilization) and development (e.g., migration of neurons or lymphocytes) as well as in normal function and health (e.g., migration of leukocytes during injury or infection).[2] In addition, it has been recognized that mechanisms that allow chemotaxis in animals can be subverted during cancer metastasis.[3] The aberrant chemotaxis of leukocytes and lymphocytes also contribute to inflammatory diseases such as atherosclerosis, asthma, and arthritis.[4][5][6][7] Sub-cellular components, such as the polarity patch generated by mating yeast, may also display chemotactic behavior.[8]
Positive chemotaxis occurs if the movement is toward a higher concentration of the chemical in question; negative chemotaxis if the movement is in the opposite direction. Chemically prompted kinesis (randomly directed or nondirectional) can be called chemokinesis.
History of chemotaxis research[edit]
Although migration of cells was detected from the early days of the development of microscopy by Leeuwenhoek, a Caltech lecture regarding chemotaxis propounds that 'erudite description of chemotaxis was only first made by T. W. Engelmann (1881) and W. F. Pfeffer (1884) in bacteria, and H. S. Jennings (1906) in ciliates'.[9] The Nobel Prize laureate I. Metchnikoff also contributed to the study of the field during 1882 to 1886, with investigations of the process as an initial step of phagocytosis.[10] The significance of chemotaxis in biology and clinical pathology was widely accepted in the 1930s, and the most fundamental definitions underlying the phenomenon were drafted by this time.[by whom?] The most important aspects in quality control of chemotaxis assays were described by H. Harris in the 1950s.[11] In the 1960s and 1970s, the revolution of modern cell biology and biochemistry provided a series of novel techniques that became available to investigate the migratory responder cells and subcellular fractions responsible for chemotactic activity.[12] The availability of this technology led to the discovery of C5a, a major chemotactic factor involved in acute inflammation. The pioneering works of J. Adler modernized Pfeffer's capillary assay and represented a significant turning point in understanding the whole process of intracellular signal transduction of bacteria.[13][14]
Bacterial chemotaxis—general characteristics[edit]

Some bacteria, such as E. coli, have several flagella per cell (4–10 typically). These can rotate in two ways:
- Counter-clockwise rotation aligns the flagella into a single rotating bundle, causing the bacterium to swim in a straight line; and
- Clockwise rotation breaks the flagella bundle apart such that each flagellum points in a different direction, causing the bacterium to tumble in place.[15]
The directions of rotation are given for an observer outside the cell looking down the flagella toward the cell.[16]
Behavior[edit]
The overall movement of a bacterium is the result of alternating tumble and swim phases, called run-and-tumble motion.[17] As a result, the trajectory of a bacterium swimming in a uniform environment will form a random walk with relatively straight swims interrupted by random tumbles that reorient the bacterium.[18] Bacteria such as E. coli are unable to choose the direction in which they swim, and are unable to swim in a straight line for more than a few seconds due to rotational diffusion; in other words, bacteria "forget" the direction in which they are going. By repeatedly evaluating their course, and adjusting if they are moving in the wrong direction, bacteria can direct their random walk motion toward favorable locations.[19]
In the presence of a chemical gradient bacteria will chemotax, or direct their overall motion based on the gradient. If the bacterium senses that it is moving in the correct direction (toward attractant/away from repellent), it will keep swimming in a straight line for a longer time before tumbling; however, if it is moving in the wrong direction, it will tumble sooner. Bacteria like E. coli use temporal sensing to decide whether their situation is improving or not, and in this way, find the location with the highest concentration of attractant, detecting even small differences in concentration.[20]
This biased random walk is a result of simply choosing between two methods of random movement; namely tumbling and straight swimming.[21] The helical nature of the individual flagellar filament is critical for this movement to occur. The protein structure that makes up the flagellar filament, flagellin, is conserved among all flagellated bacteria.[22] Vertebrates seem to have taken advantage of this fact by possessing an immune receptor (TLR5) designed to recognize this conserved protein. [23]
As in many instances in biology, there are bacteria that do not follow this rule. Many bacteria, such as Vibrio, are monoflagellated and have a single flagellum at one pole of the cell. Their method of chemotaxis is different. Others possess a single flagellum that is kept inside the cell wall. These bacteria move by spinning the whole cell, which is shaped like a corkscrew.[24][page needed]
Signal transduction[edit]

Chemical gradients are sensed through multiple transmembrane receptors, called methyl-accepting chemotaxis proteins (MCPs), which vary in the molecules that they detect.[25] Thousands of MCP receptors are known to be encoded across the bacterial kingdom.[26] These receptors may bind attractants or repellents directly or indirectly through interaction with proteins of periplasmatic space.[27] The signals from these receptors are transmitted across the plasma membrane into the cytosol, where Che proteins are activated.[28] The Che proteins alter the tumbling frequency, and alter the receptors.[28]
Flagellum regulation[edit]
The proteins CheW and CheA bind to the receptor. The absence of receptor activation results in autophosphorylation in the histidine kinase, CheA, at a single highly conserved histidine residue.[29][better source needed] CheA, in turn, transfers phosphoryl groups to conserved aspartate residues in the response regulators CheB and CheY; CheA is a histidine kinase and it does not actively transfer the phosphoryl group, rather, the response regulator CheB takes the phosphoryl group from CheA.[citation needed] This mechanism of signal transduction is called a two-component system, and it is a common form of signal transduction in bacteria.[citation needed] CheY induces tumbling by interacting with the flagellar switch protein FliM, inducing a change from counter-clockwise to clockwise rotation of the flagellum. Change in the rotation state of a single flagellum can disrupt the entire flagella bundle and cause a tumble.[citation needed]
Receptor regulation[edit]

CheB, when activated by CheA, acts as a methylesterase, removing methyl groups from glutamate residues on the cytosolic side of the receptor; it works antagonistically with CheR, a methyltransferase, which adds methyl residues to the same glutamate residues.[30] If the level of an attractant remains high, the level of phosphorylation of CheA (and, therefore, CheY and CheB) will remain low, the cell will swim smoothly, and the level of methylation of the MCPs will increase (because CheB-P is not present to demethylate).[30] The MCPs no longer respond to the attractant when they are fully methylated; therefore, even though the level of attractant might remain high, the level of CheA-P (and CheB-P) increases and the cell begins to tumble.[30] The MCPs can be demethylated by CheB-P, and, when this happens, the receptors can once again respond to attractants.[30] The situation is the opposite with regard to repellents: fully methylated MCPs respond best to repellents, while least-methylated MCPs respond worst to repellents.[citation needed] This regulation allows the bacterium to 'remember' chemical concentrations from the recent past, a few seconds, and compare them to those it is currently experiencing, thus 'know' whether it is traveling up or down a gradient. [31] that bacteria have to chemical gradients, other mechanisms are involved in increasing the absolute value of the sensitivity on a given background. Well-established examples are the ultra-sensitive response of the motor to the CheY-P signal, and the clustering of chemoreceptors.[32][33]
Chemoattractants and chemorepellents[edit]
Chemoattractants and chemorepellents are inorganic or organic substances possessing chemotaxis-inducer effect in motile cells. These chemotactic ligands create chemical concentration gradients that organisms, prokaryotic and eukaryotic, move toward or away from, respectively.[34]
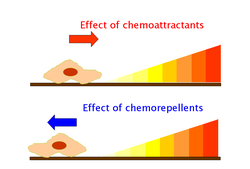
Effects of chemoattractants are elicited via chemoreceptors such as methyl-accepting chemotaxis proteins (MCP).[35] MCPs in E.coli include Tar, Tsr, Trg and Tap.[36] Chemoattracttants to Trg include ribose and galactose with phenol as a chemorepellent. Tap and Tsr recognize dipeptides and serine as chemoattractants, respectively.[36]
Chemoattractants or chemorepellents bind MCPs at its extracellular domain; an intracellular signaling domain relays the changes in concentration of these chemotactic ligands to downstream proteins like that of CheA which then relays this signal to flagellar motors via phosphorylated CheY (CheY-P).[35] CheY-P can then control flagellar rotation influencing the direction of cell motility.[35]
For E.coli, S. meliloti, and R. spheroides, the binding of chemoattractants to MCPs inhibit CheA and therefore CheY-P activity, resulting in smooth runs, but for B. substilis, CheA activity increases.[35] Methylation events in E.coli cause MCPs to have lower affinity to chemoattractants which causes increased activity of CheA and CheY-P resulting in tumbles.[35] In this way cells are able to adapt to the immediate chemoattractant concentration and detect further changes to modulate cell motility.[35]
Chemoattractants in eukaryotes are well characterized for immune cells. Formyl peptides, such as fMLF, attract leukocytes such as neutrophils and macrophages, causing movement toward infection sites.[37] Non-acylated methioninyl peptides do not act as chemoattractants to neutrophils and macrophages.[37] Leukocytes also move toward chemoattractants C5a, a complement component, and pathogen-specific ligands on bacteria.[37]
Mechanisms concerning chemorepellents are less known than chemoattractants. Although chemorepellents work to confer an avoidance response in organisms, Tetrahymena thermophila adapt to a chemorepellent, Netrin-1 peptide, within 10 minutes of exposure; however, exposure to chemorepellents such as GTP, PACAP-38, and nociceptin show no such adaptations.[38] GTP and ATP are chemorepellents in micro-molar concentrations to both Tetrahymena and Paramecium. These organisms avoid these molecules by producing avoiding reactions to re-orient themselves away from the gradient.[39]
Eukaryotic chemotaxis[edit]

The mechanism of chemotaxis that eukaryotic cells employ is quite different from that in the bacteria E. coli; however, sensing of chemical gradients is still a crucial step in the process.[40][better source needed] Due to their small size and other biophysical constraints, E. coli cannot directly detect a concentration gradient.[41] Instead, they employ temporal gradient sensing, where they move over larger distances several times their own width and measure the rate at which perceived chemical concentration changes.[42][43]
Eukaryotic cells are much larger than prokaryotes and have receptors embedded uniformly throughout the cell membrane.[42] Eukaryotic chemotaxis involves detecting a concentration gradient spatially by comparing the asymmetric activation of these receptors at the different ends of the cell.[42] Activation of these receptors results in migration towards chemoattractants, or away from chemorepellants.[42] In mating yeast, which are non-motile, patches of polarity proteins on the cell cortex can relocate in a chemotactic fashion up pheromone gradients.[44][8]
It has also been shown that both prokaryotic and eukaryotic cells are capable of chemotactic memory.[43][45] In prokaryotes, this mechanism involves the methylation of receptors called methyl-accepting chemotaxis proteins (MCPs).[43] This results in their desensitization and allows prokaryotes to "remember" and adapt to a chemical gradient.[43] In contrast, chemotactic memory in eukaryotes can be explained by the Local Excitation Global Inhibition (LEGI) model.[45][46] LEGI involves the balance between a fast excitation and delayed inhibition which controls downstream signaling such as Ras activation and PIP3 production.[47]
Levels of receptors, intracellular signalling pathways and the effector mechanisms all represent diverse, eukaryotic-type components. In eukaryotic unicellular cells, amoeboid movement and cilium or the eukaryotic flagellum are the main effectors (e.g., Amoeba or Tetrahymena).[48][49] Some eukaryotic cells of higher vertebrate origin, such as immune cells also move to where they need to be. Besides immune competent cells (granulocyte, monocyte, lymphocyte) a large group of cells—considered previously to be fixed into tissues—are also motile in special physiological (e.g., mast cell, fibroblast, endothelial cells) or pathological conditions (e.g., metastases).[50] Chemotaxis has high significance in the early phases of embryogenesis as development of germ layers is guided by gradients of signal molecules.[51][52]
Motility[edit]
Unlike motility in bacterial chemotaxis, the mechanism by which eukaryotic cells physically move is unclear. There appear to be mechanisms by which an external chemotactic gradient is sensed and turned into an intracellular PIP3 gradient, which results in a gradient and the activation of a signaling pathway, culminating in the polymerisation of actin filaments. The growing distal end of actin filaments develops connections with the internal surface of the plasma membrane via different sets of peptides and results in the formation of anterior pseudopods and posterior uropods. Cilia of eukaryotic cells can also produce chemotaxis; in this case, it is mainly a Ca2+-dependent induction of the microtubular system of the basal body and the beat of the 9 + 2 microtubules within cilia. The orchestrated beating of hundreds of cilia is synchronized by a submembranous system built between basal bodies. The details of the signaling pathways are still not totally clear.
[edit]

Chemotaxis refers to the directional migration of cells in response to chemical gradients; several variations of chemical-induced migration exist as listed below.
- Chemokinesis refers to an increase in cellular motility in response to chemicals in the surrounding environment. Unlike chemotaxis, the migration stimulated by chemokinesis lacks directionality, and instead increases environmental scanning behaviors.[53]
- In haptotaxis the gradient of the chemoattractant is expressed or bound on a surface, in contrast to the classical model of chemotaxis, in which the gradient develops in a soluble fluid.[54] The most common biologically active haptotactic surface is the extracellular matrix (ECM); the presence of bound ligands is responsible for induction of transendothelial migration and angiogenesis.
- Necrotaxis embodies a special type of chemotaxis when the chemoattractant molecules are released from necrotic or apoptotic cells. Depending on the chemical character of released substances, necrotaxis can accumulate or repel cells, which underlines the pathophysiological significance of this phenomenon.
Receptors[edit]
In general, eukaryotic cells sense the presence of chemotactic stimuli through the use of 7-transmembrane (or serpentine) heterotrimeric G-protein-coupled receptors, a class representing a significant portion of the genome.[55] Some members of this gene superfamily are used in eyesight (rhodopsins) as well as in olfaction (smelling).[56][57] The main classes of chemotaxis receptors are triggered by:
- Formyl peptides - formyl peptide receptors (FPR),
- Chemokines - chemokine receptors (CCR or CXCR), and
- Leukotrienes - leukotriene receptors (BLT).[58]
However, induction of a wide set of membrane receptors (e.g., cyclic nucleotides, amino acids, insulin, vasoactive peptides) also elicit migration of the cell.[59]
Chemotactic selection[edit]

While some chemotaxis receptors are expressed in the surface membrane with long-term characteristics, as they are determined genetically, others have short-term dynamics, as they are assembled ad hoc in the presence of the ligand.[60] The diverse features of the chemotaxis receptors and ligands allows for the possibility of selecting chemotactic responder cells with a simple chemotaxis assay By chemotactic selection, we can determine whether a still-uncharacterized molecule acts via the long- or the short-term receptor pathway.[61] The term chemotactic selection is also used to designate a technique that separates eukaryotic or prokaryotic cells according to their chemotactic responsiveness to selector ligands.[62][non-primary source needed][non-primary source needed]
Chemotactic ligands[edit]


The number of molecules capable of eliciting chemotactic responses is relatively high, and we can distinguish primary and secondary chemotactic molecules.[citation needed] The main groups of the primary ligands are as follows:
- Formyl peptides are di-, tri-, tetrapeptides of bacterial origin, formylated on the N-terminus of the peptide.[citation needed][63] They are released from bacteria in vivo or after decomposition of the cell[ a typical member of this group is the N-formylmethionyl-leucyl-phenylalanine (abbreviated fMLF or fMLP).[citation needed] Bacterial fMLF is a key component of inflammation has characteristic chemoattractant effects in neutrophil granulocytes and monocytes.[citation needed] The chemotactic factor ligands and receptors related to formyl peptides are summarized in the related article, Formyl peptide receptors.
- Complement 3a (C3a) and complement 5a (C5a) are intermediate products of the complement cascade.[citation needed] Their synthesis is joined to the three alternative pathways (classical, lectin-dependent, and alternative) of complement activation by a convertase enzyme.[citation needed] The main target cells of these derivatives are neutrophil granulocytes and monocytes as well.[citation needed]
- Chemokines belong to a special class of cytokines; not only do their groups (C, CC, CXC, CX3C chemokines) represent structurally related molecules with a special arrangement of disulfide bridges but also their target cell specificity is diverse.[citation needed] CC chemokines act on monocytes (e.g., RANTES), and CXC chemokines are neutrophil granulocyte-specific (e.g., IL-8).[citation needed] Investigations of the three-dimensional structures of chemokines provided evidence that a characteristic composition of beta-sheets and an alpha helix provides expression of sequences required for interaction with the chemokine receptors.[citation needed] Formation of dimers and their increased biological activity was demonstrated by crystallography of several chemokines, e.g. IL-8.[citation needed]
- Metabolites of polyunsaturated fatty acids
- Leukotrienes are eicosanoid lipid mediators made by the metabolism of arachidonic acid by ALOX5 (also termed 5-lipoxygenase). Their most prominent member with chemotactic factor activity is leukotriene B4, which elicits adhesion, chemotaxis, and aggregation of leukocytes. The chemoattractant action of LTB4 is induced via either of two G protein–coupled receptors, BLT1 and BLT2, which are highly expressed in cells involved in inflammation and allergy.[64]
- The family of 5-Hydroxyicosatetraenoic acid eicosanoids are arachidonic acid metabolites also formed by ALOX5. Three members of the family form naturally and have prominent chemotactic activity. These, listed in order of decreasing potency, are: 5-oxo-eicosatetraenoic acid, 5-oxo-15-hydroxy-eicosatetraenoic acid, and 5-Hydroxyeicosatetraenoic acid. This family of agonists stimulates chemotactic responses in human eosinophils, neutrophils, and monocytes by binding to the Oxoeicosanoid receptor 1, which like the receptors for leukotriene B4, is a G protein-coupled receptor.[64] Aside from the skin, neutrophils are the body's first line of defense against bacterial infections. After leaving nearby blood vessels, these cells recognize chemicals produced by bacteria in a cut or scratch and migrate "toward the smell".
- 5-hydroxyeicosatrienoic acid and 5-oxoeicosatrienoic acid are metabolites of Mead acid (5Z,8Z,11Z-eicosatrirenoid acid); they stimulate leukocyte chemotaxis through the oxoeicosanoid receptor 1[65] with 5-oxoeicosatrienoic acid being as potent as its arachidonic acid-derived analog, 5-oxo-eicosatetraenoic acid, in stimulating human blood eosinophil and neutrophil chemotaxis.[64]
- 12-Hydroxyeicosatetraenoic acid is an eicosanoid metabolite of arachidonic acid made by ALOX12 which stimulates leukocyte chemotaxis through the leukotriene B4 receptor, BLT2.[64]
- Prostaglandin D2 is an eicosanoid metabolite of arachidononic acid made by cyclooxygenase 1 or cyclooxygenase 2 that stimulates chemotaxis through the Prostaglandin DP2 receptor. It elicits chemotactic responses in eosinophils, basophils, and T helper cells of the Th2 subtype.[66][non-primary source needed][non-primary source needed]
- 12-Hydroxyheptadecatrienoic acid is a non-eicosanoid metabolite of arachidonic acid made by cyclooxygenase 1 or cyclooxygenase 2 that stimulates leukocyte chemotaxis though the leukotriene B4 receptor, BLT2.[67][non-primary source needed][non-primary source needed]
- 15-oxo-eicosatetraenoic acid is an eicosanoid metabolite of arachidonic acid made my ALOX15; it has weak chemotactic activity for human monocytes (sees 15-Hydroxyeicosatetraenoic acid#15-oxo-ETE).[68][non-primary source needed][non-primary source needed] The receptor or other mechanism by which this metabolite stimulates chemotaxis has not been elucidated.
Chemotactic range fitting[edit]

Chemotactic responses elicited by ligand-receptor interactions vary with the concentration of the ligand. Investigations of ligand families (e.g. amino acids or oligopeptides) demonstrates that chemoattractant activity occurs over a wide range, while chemorepellent activities have narrow ranges.[69]
Clinical significance[edit]
A changed migratory potential of cells has relatively high importance in the development of several clinical symptoms and syndromes. Altered chemotactic activity of extracellular (e.g., Escherichia coli) or intracellular (e.g., Listeria monocytogenes) pathogens itself represents a significant clinical target. Modification of endogenous chemotactic ability of these microorganisms by pharmaceutical agents can decrease or inhibit the ratio of infections or spreading of infectious diseases. Apart from infections, there are some other diseases wherein impaired chemotaxis is the primary etiological factor, as in Chédiak–Higashi syndrome, where giant intracellular vesicles inhibit normal migration of cells.
| Type of disease | Chemotaxis increased | Chemotaxis decreased |
|---|---|---|
| Infections | Inflammations | AIDS, Brucellosis |
| Chemotaxis results in the disease | — | Chédiak–Higashi syndrome, Kartagener syndrome |
| Chemotaxis is affected | Atherosclerosis, arthritis, periodontitis, psoriasis, reperfusion injury, metastatic tumors | Multiple sclerosis, Hodgkin disease, male infertility |
| Intoxications | Asbestos, benzpyrene | Hg and Cr salts, ozone |
Mathematical models[edit]
Several mathematical models of chemotaxis were developed depending on the type of
- Migration (e.g., basic differences of bacterial swimming, movement of unicellular eukaryotes with cilia/flagellum and amoeboid migration)
- Physico-chemical characteristics of the chemicals (e.g., diffusion) working as ligands
- Biological characteristics of the ligands (attractant, neutral, and repellent molecules)
- Assay systems applied to evaluate chemotaxis (see incubation times, development, and stability of concentration gradients)
- Other environmental effects possessing direct or indirect influence on the migration (lighting, temperature, magnetic fields, etc.)
Although interactions of the factors listed above make the behavior of the solutions of mathematical models of chemotaxis rather complex, it is possible to describe the basic phenomenon of chemotaxis-driven motion in a straightforward way. Indeed, let us denote with the spatially non-uniform concentration of the chemo-attractant and as its gradient. Then the chemotactic cellular flow (also called current) that is generated by the chemotaxis is linked to the above gradient by the law:[70]
where is the spatial density of the cells and is the so-called 'Chemotactic coefficient' - is often not constant, but a decreasing function of the chemo-attractant. For some quantity that is subject to total flux and generation/destruction term , it is possible to formulate a continuity equation:
where is the divergence. This general equation applies to both the cell density and the chemo-attractant. Therefore, incorporating a diffusion flux into the total flux term, the interactions between these quantities are governed by a set of coupled reaction-diffusion partial differential equations describing the change in and :[70]
where describes the growth in cell density, is the kinetics/source term for the chemo-attractant, and the diffusion coefficients for cell density and the chemo-attractant are respectively and .
Spatial ecology of soil microorganisms is a function of their chemotactic sensitivities towards substrate and fellow organisms.[71][non-primary source needed][non-primary source needed] The chemotactic behavior of the bacteria was proven to lead to non-trivial population patterns even in the absence of environmental heterogeneities. The presence of structural pore scale heterogeneities has an extra impact on the emerging bacterial patterns.
Measurement of chemotaxis[edit]
A wide range of techniques is available to evaluate chemotactic activity of cells or the chemoattractant and chemorepellent character of ligands. The basic requirements of the measurement are as follows:
- Concentration gradients can develop relatively quickly and persist for a long time in the system
- Chemotactic and chemokinetic activities are distinguished
- Migration of cells is free toward and away on the axis of the concentration gradient
- Detected responses are the results of active migration of cells
Despite the fact that an ideal chemotaxis assay is still not available, there are several protocols and pieces of equipment that offer good correspondence with the conditions described above. The most commonly used are summarised in the table below:
| Type of assay | Agar-plate assays | Two-chamber assays | Others |
|---|---|---|---|
| Examples |
|
|
|
Artificial chemotactic systems[edit]
Chemical robots that use artificial chemotaxis to navigate autonomously have been designed.[72][73] Applications include targeted delivery of drugs in the body.[74] More recently, enzyme molecules have also shown positive chemotactic behavior in the gradient of their substrates.[75] The thermodynamically-favorable binding of enzymes to their specific substrates is recognized as the origin of enzymatic chemotaxis.[76] Additionally, enzymes in cascades have also shown substrate-driven chemotactic aggregation.[77]
Apart from active enzymes, non-reacting molecules also show chemotactic behavior. This has been demonstrated by using dye molecules that move directionally in gradients of polymer solution through favorable hydrophobic interactions.[78]
See also[edit]
References[edit]
- ^ Chisholm, Hugh, ed. (1911). . Encyclopædia Britannica. Vol. 6 (11th ed.). Cambridge University Press. p. 77.
- ^ de Oliveira S, Rosowski EE, Huttenlocher A (May 2016). "Neutrophil migration in infection and wound repair: going forward in reverse". Nature Reviews. Immunology. 16 (6): 378–91. doi:10.1038/nri.2016.49. PMC 5367630. PMID 27231052.
- ^ Stuelten CH, Parent CA, Montell DJ (May 2018). "Cell motility in cancer invasion and metastasis: insights from simple model organisms". Nature Reviews. Cancer. 18 (5): 296–312. doi:10.1038/nrc.2018.15. PMC 6790333. PMID 29546880.
- ^ Li J, Ley K (January 2015). "Lymphocyte migration into atherosclerotic plaque". Arteriosclerosis, Thrombosis, and Vascular Biology. 35 (1): 40–9. doi:10.1161/ATVBAHA.114.303227. PMC 4429868. PMID 25301842.
- ^ Gelfand EW (October 2017). "Importance of the leukotriene B4-BLT1 and LTB4-BLT2 pathways in asthma". Seminars in Immunology. 33: 44–51. doi:10.1016/j.smim.2017.08.005. PMC 5679233. PMID 29042028.
- ^ Planagumà A, Domènech T, Pont M, Calama E, García-González V, López R, Aulí M, López M, Fonquerna S, Ramos I, de Alba J, Nueda A, Prats N, Segarra V, Miralpeix M, Lehner MD (October 2015). "Combined anti CXC receptors 1 and 2 therapy is a promising anti-inflammatory treatment for respiratory diseases by reducing neutrophil migration and activation". Pulmonary Pharmacology & Therapeutics. 34: 37–45. doi:10.1016/j.pupt.2015.08.002. PMID 26271598.
- ^ Rana AK, Li Y, Dang Q, Yang F (December 2018). "Monocytes in rheumatoid arthritis: Circulating precursors of macrophages and osteoclasts and, their heterogeneity and plasticity role in RA pathogenesis". International Immunopharmacology. 65: 348–359. doi:10.1016/j.intimp.2018.10.016. PMID 30366278. S2CID 53116963.
- ^ a b Ghose, Debraj; Jacobs, Katherine; Ramirez, Samuel; Elston, Timothy; Lew, Daniel (1 June 2021). "Chemotactic movement of a polarity site enables yeast cells to find their mates". Proceedings of the National Academy of Sciences. 118 (22): e2025445118. Bibcode:2021PNAS..11825445G. doi:10.1073/pnas.2025445118. ISSN 0027-8424. PMC 8179161. PMID 34050026.
- ^ Chemotaxis Lecture. Uploaded in 2007. available at: http://www.rpgroup.caltech.edu/courses/aph161/2007/lectures/ChemotaxisLecture.pdf Archived 2010-06-19 at the Wayback Machine (Last inspected: 15/04/17)
- ^ Élie Metchnikoff". Encyclopædia Britannica. Encyclopædia Britannica, Inc.
- ^ Keller-Segel Models for Chemotaxis. 2012. available at: http://www.isn.ucsd.edu/courses/Beng221/problems/2012/BENG221_Project%20-%20Roberts%20Chung%20Yu%20Li.pdf Archived 29 August 2017 at the Wayback Machine (Last inspected by April 2017)
- ^ Snyderman R, Gewurz H, Mergenhagen SE (August 1968). "Interactions of the complement system with endotoxic lipopolysaccharide. Generation of a factor chemotactic for polymorphonuclear leukocytes". The Journal of Experimental Medicine. 128 (2): 259–75. doi:10.1084/jem.128.2.259. PMC 2138524. PMID 4873021.
- ^ Adler J, Tso WW (June 1974). ""Decision"-making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli". Science. 184 (4143): 1292–4. Bibcode:1974Sci...184.1292A. doi:10.1126/science.184.4143.1292. PMID 4598187. S2CID 7221477.
- ^ Berg, Howard (2004). Berg, Howard C (ed.). E. coli in Motion. Biological and Medical Physics, Biomedical Engineering. Springer. p. 15, 19–29. doi:10.1007/b97370. ISBN 0-387-00888-8. S2CID 35733036.
- ^ Yuan J, Fahrner KA, Turner L, Berg HC (July 2010). "Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor". Proceedings of the National Academy of Sciences of the United States of America. 107 (29): 12846–9. Bibcode:2010PNAS..10712846Y. doi:10.1073/pnas.1007333107. PMC 2919929. PMID 20615986.
- ^ "Bacterial Chemotaxis" (PDF). Archived (PDF) from the original on 6 May 2017.
- ^ Berg HC, Brown DA (October 1972). "Chemotaxis in Escherichia coli analysed by Three-dimensional Tracking". Nature. 239 (5374): 500–504. Bibcode:1972Natur.239..500B. doi:10.1038/239500a0. PMID 4563019. S2CID 1909173.
- ^ Sourjik V, Wingreen NS (April 2012). "Responding to chemical gradients: bacterial chemotaxis". Current Opinion in Cell Biology. 24 (2): 262–268. doi:10.1016/j.ceb.2011.11.008. PMC 3320702. PMID 22169400.
- ^ Berg, Howard C. (1993). Random walks in biology (Expanded, rev. ed.). Princeton, NJ: Princeton Univ. Press. pp. 83–94. ISBN 978-0-691-00064-0.
- ^ Sourjik V, Wingreen N (April 2012). "Responding to Chemical Gradients: Bacterial Chemotaxis". Current Opinion in Cell Biology. 24 (2): 262–8. doi:10.1016/j.ceb.2011.11.008. PMC 3320702. PMID 22169400.
- ^ Macnab RM, Koshland DE (September 1972). "The gradient-sensing mechanism in bacterial chemotaxis". Proceedings of the National Academy of Sciences of the United States of America. 69 (9): 2509–12. Bibcode:1972PNAS...69.2509M. doi:10.1073/pnas.69.9.2509. PMC 426976. PMID 4560688.
- ^ Nedeljković, Marko; Sastre, Diego; Sundberg, Eric (14 July 2021). "Bacterial Flagellar Filament: A Supramolecular Multifunctional Nanostructure". International Journal of Molecular Sciences. 22 (14): 7521. doi:10.3390/ijms22147521. PMC 8306008. PMID 34299141.
- ^ Zhong, Maohua; Yan, Huimin; Li, Yaoming (October 2017). "Flagellin: a unique microbe-associated molecular pattern and a multi-faceted immunomodulator". Cellular & Molecular Immunology. 14 (10): 862–864. doi:10.1038/cmi.2017.78. ISSN 2042-0226. PMC 5649114. PMID 28845044.
- ^ Berg HC (2003). E. coli in motion. New York, NY: Springer. ISBN 978-0-387-00888-2.[page needed]
- ^ Wadhams, George H.; Armitage, Judith P. (December 2004). "Making sense of it all: bacterial chemotaxis". Nature Reviews Molecular Cell Biology. 5 (12): 1024–1037. doi:10.1038/nrm1524. PMID 15573139. S2CID 205493118.
- ^ Galperin, Michael (June 2005). "A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts". BMC Microbiology. 5: 35. doi:10.1186/1471-2180-5-35. PMC 1183210. PMID 15955239.
- ^ Gennaro Auletta (2011). Cognitive Biology: Dealing with Information from Bacteria to Minds. United States: Oxford University Press. p. 266. ISBN 978-0-19-960848-5.
- ^ a b Falke, Joseph J.; Bass, Randal B.; Butler, Scott L.; Chervitz, Stephen A.; Danielson, Mark A. (1997). "THE TWO-COMPONENT SIGNALING PATHWAY OF BACTERIAL CHEMOTAXIS: A Molecular View of Signal Transduction by Receptors, Kinases, and Adaptation Enzymes". Annual Review of Cell and Developmental Biology. 13: 457–512. doi:10.1146/annurev.cellbio.13.1.457. ISSN 1081-0706. PMC 2899694. PMID 9442881.
- ^ ToxCafe (2 June 2011). "Chemotaxis". Archived from the original on 11 July 2015. Retrieved 23 March 2017 – via YouTube.
- ^ a b c d Wadhams, George H.; Armitage, Judith P. (December 2004). "Making sense of it all: bacterial chemotaxis". Nature Reviews Molecular Cell Biology. 5 (12): 1024–1037. doi:10.1038/nrm1524. ISSN 1471-0080. PMID 15573139. S2CID 205493118.
- ^ Shu Chien; Peter C Y Chen; Y C Fung (2008). An Introductory Text to Bioengineering (Advanced Series in Biomechanics - Vol. 4). Singapore: World Scientific Publishing Co. Pte. Ltd. p. 418. ISBN 9789812707932.
- ^ Cluzel P, Surette M, Leibler S (March 2000). "An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells". Science. 287 (5458): 1652–5. Bibcode:2000Sci...287.1652C. doi:10.1126/science.287.5458.1652. PMID 10698740. S2CID 5334523.
- ^ Sourjik V (December 2004). "Receptor clustering and signal processing in E. coli chemotaxis". Trends in Microbiology. 12 (12): 569–76. CiteSeerX 10.1.1.318.4824. doi:10.1016/j.tim.2004.10.003. PMID 15539117.
- ^ Xu F, Bierman R, Healy F, Nguyen H (2016). "A multi-scale model of Escherichia coli chemotaxis from intracellular signaling pathway to motility and nutrient uptake in nutrient gradient and isotropic fluid environments". Computers & Mathematics with Applications. 71 (11): 2466–2478. doi:10.1016/j.camwa.2015.12.019.
- ^ a b c d e f Szurmant H, Ordal GW (June 2004). "Diversity in chemotaxis mechanisms among the bacteria and archaea". Microbiology and Molecular Biology Reviews. 68 (2): 301–19. doi:10.1128/MMBR.68.2.301-319.2004. PMC 419924. PMID 15187186.
- ^ a b Yamamoto K, Macnab RM, Imae Y (January 1990). "Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli". Journal of Bacteriology. 172 (1): 383–8. doi:10.1128/jb.172.1.383-388.1990. PMC 208443. PMID 2403544.
- ^ a b c Schiffmann E, Corcoran BA, Wahl SM (March 1975). "N-formylmethionyl peptides as chemoattractants for leucocytes". Proceedings of the National Academy of Sciences of the United States of America. 72 (3): 1059–62. Bibcode:1975PNAS...72.1059S. doi:10.1073/pnas.72.3.1059. PMC 432465. PMID 1093163.
- ^ Kuruvilla H, Schmidt B, Song S, Bhajjan M, Merical M, Alley C, Griffin C, Yoder D, Hein J, Kohl D, Puffenberger C, Petroff D, Newcomer E, Good K, Heston G, Hurtubise A (2016). "Netrin-1 Peptide Is a Chemorepellent in Tetrahymena thermophila". International Journal of Peptides. 2016: 7142868. doi:10.1155/2016/7142868. PMC 4830718. PMID 27123011.
- ^ Hennessey TM (June 2005). "Responses of the ciliates Tetrahymena and Paramecium to external ATP and GTP". Purinergic Signalling. 1 (2): 101–10. doi:10.1007/s11302-005-6213-1. PMC 2096533. PMID 18404496.
- ^ Köhidai L (2016), "Chemotaxis as an Expression of Communication of Tetrahymena", in Witzany G, Nowacki M (eds.), Biocommunication of Ciliates, pp. 65–82, doi:10.1007/978-3-319-32211-7_5, ISBN 978-3-319-32211-7
- ^ Berg, H.C.; Purcell, E.M. (November 1977). "Physics of chemoreception". Biophysical Journal. 20 (2): 193–219. Bibcode:1977BpJ....20..193B. doi:10.1016/s0006-3495(77)85544-6. ISSN 0006-3495. PMC 1473391. PMID 911982.
- ^ a b c d Levine, Herbert; Rappel, Wouter-Jan (1 February 2013). "The physics of eukaryotic chemotaxis". Physics Today. 66 (2): 24–30. Bibcode:2013PhT....66b..24L. doi:10.1063/PT.3.1884. PMC 3867297. PMID 24363460.
- ^ a b c d Vladimirov N, Sourjik V (November 2009). "Chemotaxis: how bacteria use memory". Biological Chemistry. 390 (11): 1097–104. doi:10.1515/BC.2009.130. PMID 19747082. S2CID 207440927.
- ^ Ghose, Debraj; Lew, Daniel (1 May 2020). "Mechanistic insights into actin-driven polarity site movement in yeast". Molecular Biology of the Cell. 31 (10): 1085–1102. doi:10.1091/mbc.e20-01-0040. ISSN 1059-1524. PMC 7346724. PMID 32186970.
- ^ a b Skoge M, Yue H, Erickstad M, Bae A, Levine H, Groisman A, Loomis WF, Rappel WJ (October 2014). "Cellular memory in eukaryotic chemotaxis". Proceedings of the National Academy of Sciences of the United States of America. 111 (40): 14448–53. Bibcode:2014PNAS..11114448S. doi:10.1073/pnas.1412197111. PMC 4210025. PMID 25249632.
- ^ Kutscher B, Devreotes P, Iglesias PA (February 2004). "Local excitation, global inhibition mechanism for gradient sensing: an interactive applet". Science's STKE. 2004 (219): pl3. doi:10.1126/stke.2192004pl3. PMID 14872096. S2CID 4660870.
- ^ Xiong Y, Huang CH, Iglesias PA, Devreotes PN (October 2010). "Cells navigate with a local-excitation, global-inhibition-biased excitable network". Proceedings of the National Academy of Sciences of the United States of America. 107 (40): 17079–86. Bibcode:2010PNAS..10717079X. doi:10.1073/pnas.1011271107. PMC 2951443. PMID 20864631.
- ^ Bagorda A, Parent CA (August 2008). "Eukaryotic chemotaxis at a glance". Journal of Cell Science. 121 (Pt 16): 2621–4. CiteSeerX 10.1.1.515.32. doi:10.1242/jcs.018077. PMC 7213762. PMID 18685153.
- ^ Köhidai L (1999). "Chemotaxis: the proper physiological response to evaluate phylogeny of signal molecules". Acta Biologica Hungarica. 50 (4): 375–94. doi:10.1007/BF03543060. PMID 10735174. S2CID 248703226.
- ^ Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE (September 2007). "Cell motility and cytoskeletal regulation in invasion and metastasis". Journal of Mammary Gland Biology and Neoplasia. 12 (2–3): 143–52. doi:10.1007/s10911-007-9046-4. PMID 17557195. S2CID 31704677.
- ^ Solnica-Krezel L, Sepich DS (2012). "Gastrulation: making and shaping germ layers". Annual Review of Cell and Developmental Biology. 28: 687–717. doi:10.1146/annurev-cellbio-092910-154043. PMID 22804578. S2CID 11331182.
- ^ Shellard A, Mayor R (July 2016). "Chemotaxis during neural crest migration". Seminars in Cell & Developmental Biology. 55: 111–8. doi:10.1016/j.semcdb.2016.01.031. PMID 26820523.
- ^ Becker EL (October 1977). "Stimulated neutrophil locomotion: chemokinesis and chemotaxis". Archives of Pathology & Laboratory Medicine. 101 (10): 509–13. PMID 199132.
- ^ Carter SB (January 1967). "Haptotaxis and the mechanism of cell motility". Nature. 213 (5073): 256–60. Bibcode:1967Natur.213..256C. doi:10.1038/213256a0. PMID 6030602. S2CID 4212997.
- ^ Kim JY, Haastert PV, Devreotes PN (April 1996). "Social senses: G-protein-coupled receptor signaling pathways in Dictyostelium discoideum". Chemistry & Biology. 3 (4): 239–43. doi:10.1016/s1074-5521(96)90103-9. PMID 8807851.
- ^ Montell, Craig (November 1999). "Visual Transduction in Drosophila". Annual Review of Cell and Developmental Biology. 15 (1): 231–268. doi:10.1146/annurev.cellbio.15.1.231. PMID 10611962. S2CID 14193715.
- ^ Antunes G, Simoes de Souza FM (2016). "Olfactory receptor signaling". G Protein-Coupled Receptors - Signaling, Trafficking and Regulation. Methods in Cell Biology. Vol. 132. pp. 127–45. doi:10.1016/bs.mcb.2015.11.003. ISBN 9780128035955. PMID 26928542.
- ^ Thomas, Monica A.; Kleist, Andrew B.; Volkman, Brian F. (2018). "Decoding the chemotactic signal". Journal of Leukocyte Biology. 104 (2): 359–374. doi:10.1002/JLB.1MR0218-044. PMC 6099250. PMID 29873835.
- ^ van Haastert PJ, De Wit RJ, Konijn TM (August 1982). "Antagonists of chemoattractants reveal separate receptors for cAMP, folic acid and pterin in Dictyostelium" (PDF). Experimental Cell Research. 140 (2): 453–6. doi:10.1016/0014-4827(82)90139-2. PMID 7117406. S2CID 27784085.
- ^ Witzany, Guenther; Nowacki, Mariusz (2016). Biocommunication of Ciliates. Springer. ISBN 978-3-319-32211-7.[page needed]
- ^ Köhidai, Laszlo (2016). "Chemotaxis as an Expression of Communication of Tetrahymena". In Witzany, Guenther; Nowacki, Mariusz (eds.). Biocommunication of Ciliates. Springer. pp. 65–82. doi:10.1007/978-3-319-32211-7_5. ISBN 978-3-319-32211-7.
- ^ Köhidai L, Csaba G (July 1998). "Chemotaxis and chemotactic selection induced with cytokines (IL-8, RANTES and TNF-alpha) in the unicellular Tetrahymena pyriformis". Cytokine. 10 (7): 481–6. doi:10.1006/cyto.1997.0328. PMID 9702410. S2CID 33755476.
- ^ Zigmond SH (November 1977). "Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors". The Journal of Cell Biology. 75 (2 Pt 1): 606–16. doi:10.1083/jcb.75.2.606. PMC 2109936. PMID 264125.
- ^ a b c d Powell WS, Rokach J (April 2015). "Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1851 (4): 340–55. doi:10.1016/j.bbalip.2014.10.008. PMC 5710736. PMID 25449650.
- ^ Powell WS, Rokach J (October 2013). "The eosinophil chemoattractant 5-oxo-ETE and the OXE receptor". Progress in Lipid Research. 52 (4): 651–65. doi:10.1016/j.plipres.2013.09.001. PMC 5710732. PMID 24056189.
- ^ Matsuoka T, Narumiya S (September 2007). "Prostaglandin receptor signaling in disease". TheScientificWorldJournal. 7: 1329–47. doi:10.1100/tsw.2007.182. PMC 5901339. PMID 17767353.
- ^ Yokomizo T (February 2015). "Two distinct leukotriene B4 receptors, BLT1 and BLT2". Journal of Biochemistry. 157 (2): 65–71. doi:10.1093/jb/mvu078. PMID 25480980.
- ^ Sozzani S, Zhou D, Locati M, Bernasconi S, Luini W, Mantovani A, O'Flaherty JT (November 1996). "Stimulating properties of 5-oxo-eicosanoids for human monocytes: synergism with monocyte chemotactic protein-1 and -3". Journal of Immunology. 157 (10): 4664–71. doi:10.4049/jimmunol.157.10.4664. PMID 8906847. S2CID 23499393.
- ^ Kohidai L, Lang O and Csaba G (2003). "Chemotactic-range-fitting of amino acids and its correlations to physicochemical parameters in Tetrahymena pyriformis - Evolutionary consequences". Cellular and Molecular Biology. 49: OL487–95. PMID 14995080.
- ^ a b Murray, James D. (2002). Mathematical Biology I: An Introduction (PDF). Interdisciplinary Applied Mathematics. Vol. 17 (3rd ed.). New York: Springer. pp. 395–417. doi:10.1007/b98868. ISBN 978-0-387-95223-9. Archived (PDF) from the original on 6 May 2022.
- ^ Gharasoo M, Centler F, Fetzer I, Thullner M (2014). "How the chemotactic characteristics of bacteria can determine their population patterns". Soil Biology and Biochemistry. 69: 346–358. doi:10.1016/j.soilbio.2013.11.019.
- ^ Mackenzie, Dana (6 March 2023). "How animals follow their nose". Knowable Magazine. Annual Reviews. doi:10.1146/knowable-030623-4. S2CID 257388244. Retrieved 13 March 2023.
- ^ Reddy, Gautam; Murthy, Venkatesh N.; Vergassola, Massimo (10 March 2022). "Olfactory Sensing and Navigation in Turbulent Environments". Annual Review of Condensed Matter Physics. 13 (1): 191–213. Bibcode:2022ARCMP..13..191R. doi:10.1146/annurev-conmatphys-031720-032754. ISSN 1947-5454. S2CID 243966350.
- ^ Lagzi, István (2013). "Chemical Robotics—Chemotactic Drug Carriers". Central European Journal of Medicine. 8 (4): 377–382. doi:10.2478/s11536-012-0130-9. S2CID 84150518.
- ^ Sengupta S, Dey KK, Muddana HS, Tabouillot T, Ibele ME, Butler PJ, Sen A (January 2013). "Enzyme molecules as nanomotors". Journal of the American Chemical Society. 135 (4): 1406–14. doi:10.1021/ja3091615. PMID 23308365.
- ^ Mohajerani, F; Zhao, X; Somasundar, A; Velegol, D; Sen, A (2018). "A Theory of Enzyme Chemotaxis: From Experiments to Modeling". Biochemistry. 57 (43): 6256–6263. arXiv:1809.02530. doi:10.1021/acs.biochem.8b00801. PMID 30251529. S2CID 52816076.
- ^ Zhao X, Palacci H, Yadav V, Spiering MM, Gilson MK, Butler PJ, Hess H, Benkovic SJ, Sen A (March 2018). "Substrate-driven chemotactic assembly in an enzyme cascade". Nature Chemistry. 10 (3): 311–317. Bibcode:2018NatCh..10..311Z. doi:10.1038/nchem.2905. PMID 29461522.
- ^ Guha R, Mohajerani F, Collins M, Ghosh S, Sen A, Velegol D (November 2017). "Chemotaxis of Molecular Dyes in Polymer Gradients in Solution". Journal of the American Chemical Society. 139 (44): 15588–15591. doi:10.1021/jacs.7b08783. PMID 29064685.
Further reading[edit]
- Alberts B, Johnson A, Lewis J, Walter P, Raff MC (2002). "Bacterial Chemotaxis Depends on a Two-Component Signaling Pathway Activated by Histidine-Kinase-associated Receptors". Molecular Biology of the Cell. Taylor & Francis Group. ISBN 978-0-8153-4069-0. Retrieved 18 September 2017.
- Bagorda A, Parent CA (August 2008). "Eukaryotic chemotaxis at a glance". Journal of Cell Science. 121 (Pt 16): 2621–4. CiteSeerX 10.1.1.515.32. doi:10.1242/jcs.018077. PMC 7213762. PMID 18685153.
- Berg HC (1993). Random walks in biology (Expanded, rev. ed.). Princeton, NJ: Princeton Univ. Press. ISBN 978-0-691-00064-0.
- Berg HC (2003). E. coli in motion. Vol. 58. New York: Springer. pp. 64–65. Bibcode:2005PhT....58b..64B. doi:10.1063/1.1897527. ISBN 978-0-387-00888-2.
{{cite book}}:|journal=ignored (help) - Dusenbery DB (2009). Living at micro scale : the unexpected physics of being small. Cambridge, Mass.: Harvard University Press. ISBN 978-0-674-03116-6.
- Eisenbach M (2004). Lengeler JW (ed.). Chemotaxis. London: Imperial College Press. ISBN 978-1-86094-413-0.
- Eisenbach, Michael (December 2011). "Bacterial Chemotaxis". Encyclopedia of Life Sciences. doi:10.1002/9780470015902.a0001251.pub3. ISBN 978-0470016176.
{{cite book}}:|journal=ignored (help) - Hazelbauer GL (13 October 2012). "Bacterial chemotaxis: the early years of molecular studies". Annual Review of Microbiology. 66 (1): 285–303. doi:10.1146/annurev-micro-092611-150120. PMC 3989901. PMID 22994495.
- Jin T, Hereld D (2016). Chemotaxis: Methods and Protocols. Humana Press. ISBN 978-1-4939-3480-5.
- Miller LD, Russell MH, Alexandre G (2009). Diversity in bacterial chemotactic responses and niche adaptation. Vol. 66. pp. 53–75. doi:10.1016/S0065-2164(08)00803-4. ISBN 9780123747884. PMID 19203648.
{{cite book}}:|journal=ignored (help) - Rao CV, Kirby JR, Arkin AP (February 2004). "Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis". PLOS Biology. 2 (2): E49. doi:10.1371/journal.pbio.0020049. PMC 340952. PMID 14966542.
- Williams AH (20 December 2010). "Chemotaxis on the move - active learning teaching tool". Journal of Microbiology & Biology Education. 11 (2): 177–8. doi:10.1128/jmbe.v11i2.216. PMC 3577161. PMID 23653726.











![{\displaystyle {\begin{aligned}{\partial C \over {\partial t}}&=f(C)+\nabla \cdot \left[D_{C}\nabla C-C\chi (\varphi )\nabla \varphi \right]\\{\partial \varphi \over {\partial t}}&=g(\varphi ,C)+\nabla \cdot (D_{\varphi }\nabla \varphi )\end{aligned}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/924b3d46d7dc2a27228a7d98cf43d035a4622c41)



