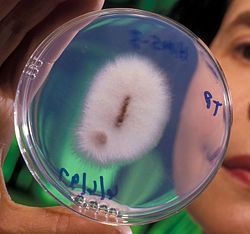
Fusarium oxysporum
| Fusarium oxysporum | |
|---|---|
| |
| Scientific classification | |
| Kingdom: | |
| Division: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | F. oxysporum
|
| Binomial name | |
| Fusarium oxysporum Schlecht. emend. Snyder & Hansen
| |
The ascomycete fungus Fusarium oxysporum Schlecht. as emended by Snyder and Hansen[1] comprises all the species, varieties and forms recognized by Wollenweber and Reinking[2] within an infrageneric grouping called section Elegans. While the species, as defined by Snyder and Hansen, has been widely accepted for more than 50 years,[3][4] more recent work indicates this taxon is actually a genetically heterogeneous polytypic morphospecies[5][6] whose strains represent some of the most abundant and widespread microbes of the global soil microflora,[7] although this last statement has not been proven or supported by actual data. These remarkably diverse and adaptable fungi have been found in soils ranging from the Sonoran Desert, to tropical and temperate forests, grasslands and soils of the tundra.[8] F. oxysporum strains are ubiquitous soil inhabitants that have the ability to exist as saprophytes, and degrade lignin[9][10] and complex carbohydrates[11][12][13] associated with soil debris. They are also pervasive plant endophytes that can colonize plant roots[14][15] and may even protect plants or be the basis of disease suppression.[16][17] Although the predominant role of these fungi in native soils may be as harmless or even beneficial plant endophytes or soil saprophytes, many strains within the F. oxysporum complex are pathogenic to plants, especially in agricultural settings.
Because the hosts of a given forma specialis usually are closely related, many have assumed that members of a forma specialis are also closely related and have descended from common ancestor.[18] However, results from recent research conducted on *Fusarium oxysporum f. sp. cubense have forced scientists to question these assumptions. Researchers used anonymous, single-copy restriction fragment length polymorphsims (RFLPs) to identify 10 clonal lineages from a collection of F. oxysporum f.sp. cubense from all over the world. These results showed that pathogens of banana could be closely related to other host’s pathogens, such as melon or tomato, as they are to each other. Exceptional amounts of genetic diversity within F. oxysporum f.sp. cubense also have been deduced from the high level of chromosomal polymorphisms found among strains, random amplified polymorphic DNA fingerprints, and from the number and geographic distribution of vegetative compatibility groups.[19]
Presented with the wide-ranging occurrence of F. oxysporum strains that are nonpathogenic, it is reasonable to expect that certain pathogenic forms were descended from originally nonpathogenic ancestors. Given the association of these fungi with plant roots, a form that is able to grow beyond the cortex and into the xylem could quickly take advantage of this ability and hopefully gain an advantage over those fungi that are restricted to the cortex. The progression of a fungus into the vascular tissue may elicit a response from the host immediately, successfully restricting the invader; or an otherwise ineffective or delayed response, reducing the vital water-conducing capacity to induce wilting.[20] On the other hand, the plant might be able to tolerate limited growth of the fungus within xylem vessels, preceded by an endophytic association.[21] In this case, any further changes in the host or parasite could disturb the relationship, in a way that fungal activities or a response of the host would result in the generation of certain disease symptoms.
Pathogenic strains of F. oxysporum have been studied for more than 100 years. The host range of these fungi is extremely broad, and includes animals, ranging from arthropods[22] to humans,[23] as well as plants, including a range of both gymnosperms and angiosperms. While collectively, plant pathogenic F. oxysporum strains have a broad host range, individual isolates usually cause disease only on a narrow range of plant species. This observation has led to the idea of "special form" or forma specialis in F. oxysporum. Formae speciales have been defined as "...an informal rank in Classification.....used for parasitic fungi characterized from a physiological standpoint (e.g. by the ability to cause disease in particular hosts) but scarcely or not at all from a morphological standpoint." Exhaustive host range studies have been conducted for relatively few formae speciales of F. oxysporum.[24] For more information on Fusarium oxysporum as a plant pathogen, see Fusarium wilt.
Different strains of F. oxysporum have been used in the purpose of producing nanomaterials (especially Silver nanoparticles).
Formae speciales (special forms)
- Fusarium oxysporum f.sp. albedinis
- Fusarium oxysporum f.sp. asparagi
- Fusarium oxysporum f.sp. batatas
- Fusarium oxysporum f.sp. betae
- Fusarium oxysporum f.sp. cannabis
- Fusarium oxysporum f.sp. cepae
- Fusarium oxysporum f.sp. ciceris
- Fusarium oxysporum f.sp. citri
- Fusarium oxysporum f.sp. coffea
- Fusarium oxysporum f.sp. cubense
- Fusarium oxysporum f.sp. cyclaminis
- Fusarium oxysporum f.sp. herbemontis
- Fusarium oxysporum f.sp. dianthi
- Fusarium oxysporum f.sp. gladioli
- Fusarium oxysporum f.sp. lactucae
- Fusarium oxysporum f.sp. lentis
- Fusarium oxysporum f.sp. lini
- Fusarium oxysporum f.sp. lycopersici
- Fusarium oxysporum f.sp. medicaginis
- Fusarium oxysporum f.sp. melonis
- Fusarium oxysporum f.sp. narcissi
- Fusarium oxysporum f.sp. nicotianae
- Fusarium oxysporum f.sp. niveum
- Fusarium oxysporum f.sp. palmarum
- Fusarium oxysporum f.sp. passiflorae
- Fusarium oxysporum f.sp. phaseoli
- Fusarium oxysporum f.sp. pisi
- Fusarium oxysporum f.sp. radicis-lycopersici
- Fusarium oxysporum f.sp. ricini
- Fusarium oxysporum f.sp. strigae
- Fusarium oxysporum f.sp. tuberosi
- Fusarium oxysporum f.sp. tulipae
- Fusarium oxysporum f.sp. vasinfectum
Patents relating to the management of Fusarium oxysporum
A number of recent patents specifically describe effective treatments of Fusarium oxysporum, reflecting its widespread importance as an agricultural pest.
- US 5,614,188: two strains of Bacillus in a composition of chitin and lime used to fight Fusarium in the soil.
- US 2004/136964 A1: Trichoderma asperellum mixed into container media (such as peat).
- US 4,714,614: a strain of Pseudomonas putida in combination with an iron chelating agent (such as EDTA).
- US 4988586: any of six types of bacteria that degrade fusaric acid, a toxin that damages plants and furthers infection.
- US 6100449 and WO 1996/032007 A1: a small genomic region (I2C) conferring resistance in transgenic tomatoes.
- US 2003/131376 A1: use of transgenic plants expressing enzymes capable of destroying Fusarium cell walls.
- US 4006265: spraying of crops with hydrogen peroxide to reduce the effect of contamination by Fusarium toxins.
- WO 2005/074687 A1: cure of infected plants by spraying with natamycin or other polyene antibiotics.
See also
References
- ^ Snyder, W.C. and Hansen, H.N. 1940. The species concept in Fusarium. Amer. J. Bot. 27:64-67.
- ^ Wollenweber, H.W. and Reinking, O.A. 1935. Die Fusarien, ihre Beschreibung, Schadwirkung und Bekampfung. P. Parey, Berlin. 365 pp.
- ^ Booth, C. 1971. The Genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, UK, 237 pp.
- ^ Nelson, P.E., Toussoun, T.A. and Marasas, W.F.O. 1983. Fusarium species: An illustrated manual for identification. Pennsylvania State University Press, University Park.
- ^ O'Donnell, K. and Cigelnik, E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7:103-116.
- ^ Waalwijk, C., De Koning, J.R.A., Baayen, R.P. and Gams, W. 1996. Discordant groupings of Fusarium spp. from section Elegans, Liseola and Dlaminia based on ribosomal ITS1 and ITS2 sequences. Mycologia 88:361-368.
- ^ Gordon, T.R. and Martyn, R.D. 1997. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35:111-128.
- ^ Stoner, M.F. 1981. Ecology of Fusarium in noncultivated soils. Pages 276-286 in: Fusarium: Diseases, Biology, and Taxonomy. P.E. Nelson, T.A. Toussoun and R.J. Cook, eds. The Pennsylvania State University Press, University Park.
- ^ Rodriguez, A., Perestelo, F., Carnicero, A., Regalado, V., Perez, R., De la Fuente, G. and Falcon, M.A.1996. Degradation of natural lignins and lignocellulosic substrates by soil-inhabiting fungi imperfecti. FEMS Microbiol. Ecol. 21:213-219.
- ^ Sutherland, J.B., Pometto, A.L. III and Crawford, D.L. 1983. Lignocellulose degradation by Fusarium species. Can. J. Bot. 61:1194-1198.
- ^ Christakopoulos, P., Kekos, D., Macris, B.J., Claeyssens, M. and Bhat, M.K. 1995. Purification and mode of action of a low molecular mass endo-1,4-B-D-glucanase from Fusarium oxysporum. J. Biotechnol. 39:85-93.
- ^ Christakopoulos, P., Nerinckx, W., Kekos, D., Macris, B. and Claeyssens, M. 1996. Purification and characterization of two low molecular mass alkaline xylanases from Fusarium oxysporum F3. J. Biotechnol. 51:181-180.
- ^ Snyder, W.C. and Hansen, H.N. 1940. The species concept in Fusarium. Amer. J. Bot. 27:64-67.
- ^ Gordon, T.R., Okamoto, D. and Jacobson, D.J. 1989. Colonization of muskmelon and nonsusceptible crops by Fusarium oxysporum f. sp. melonis and other species of Fusarium. Phytopathology 79:1095-1100.
- ^ Katan, J. 1971. Symptomless carriers of the tomato Fusarium wilt pathogen. Phytopathology 61:1213-1217.
- ^ Larkin, R.P., Hopkins, D.L. and Martin, F.N. 1993. Effect of successive watermelon plantings on Fusarium oxysporum and other microorganisms in soils suppressive and conducive to fusarium wilt of watermelon. Phytopathology 83:1097-1105.
- ^ Lemanceau, P., Bakker, P.A.H.M., DeKogel, W.J., Alabouvette, C. and Schippers, B. 1993. Antagonistic effect of nonpathogenic Fusarium oxysporum Fo47 and pseudobactin 358 upon pathogen Fusarium oxysporum f. sp. dianthi. Appl. Environ. Microbiol. 59:74-82.
- ^ O’Donnell K, Kistler H C, Cigelnik E, Ploetz R C. 1998. Evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. PNAS (internet) (Cited 2014 September 15). 95(5): 2044-2049. Available from: http://www.pnas.org/content/95/5/2044.full
- ^ Fourie G, Steenkamp ET, Gordon TR, and Viljoen A. 2009. Relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Applied and Environmental Microbiology (internet) (cited 2014 October 30). 75(14): 4770-4781. Available from: http://aem.asm.org/content/75/14/4770.full.pdf+html
- ^ Ploetz R. 2006. Fusarium Wilt of Banana is Caused by Several Pathogens Referred to as Fusarium oxysporum f. sp. cubense. Phytopathology (internet) (cited 2014 October 30). 96(6): 653-656. Available from: http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO-96-0653
- ^ Appel DJ, and Gordon TR. 1994. Local and regional variation in populations of Fusarium oxysporum from agricultural field soils. Phytopathology (internet) (Cited 2014 October 30) 84:786-791. Available from: http://www.apsnet.org/publications/phytopathology/backissues/Documents/1994Articles/Phyto84n08_786.PDF
- ^ Teetor-Barsch, G.H. and Roberts, D.W. 1983. Entomogenous Fusarium species. Mycopathologia 84:3-16.
- ^ Nelson, P.E., Dignani, M.C. and Anaissie, E.J. 1994. Taxonomy, biology, and clinical aspects of Fusarium species. Clin. Microbiol. Rev. 7:479-504.
- ^ Kistler, H.C. 2001. Evolution of host specificity in Fusarium oxysporum. Pages 70-82 in: Fusarium: Paul E. Nelson Memorial Symposium. B.A. Summerell, J.F. Leslie, D. Backhouse, W.L. Bryden and L.W. Burgess, eds. The American Phytopathological Society, St. Paul, MN.
Bibliography
- Hanks, G.R. (1996). "Control of Fusarium oxysporum f.sp. narcissi, the cause of narcissus basal rot, with thiabendazole and other fungicides". Crop Protection. 15 (6 September): 549–558. doi:10.1016/0261-2194(96)00023-3. Retrieved 3 December 2014.
{{cite journal}}: Invalid|ref=harv(help) - Hanks, Gordon; Carder, John (2003). "Management of basal rot - the narcissus disease". Pesticide Outlook. 14 (6): 260. doi:10.1039/B314848N. Retrieved 4 December 2014.
{{cite journal}}: Invalid|ref=harv(help)
