Transcription (biology)
Transcription is the synthesis of RNA under the direction of DNA. RNA synthesis, or transcription, is the process of transcribing DNA nucleotide sequence information into RNA sequence information. Both nucleic acid sequences use complementary language, and the information is simply transcribed, or copied, from one molecule to the other. DNA sequence is enzymatically copied by RNA polymerase to produce a complementary nucleotide RNA strand, called messenger RNA (mRNA), because it carries a genetic message from the DNA to the protein-synthesizing machinery of the cell. One significant difference between RNA and DNA sequence is the presence of U, or uracil in RNA instead of the T, or thymine of DNA. In the case of protein-encoding DNA, transcription is the first step that usually leads to the expression of the genes, by the production of the mRNA intermediate, which is a faithful transcript of the gene's protein-building instruction. The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit. A DNA transcription unit that is translated into protein contains sequences that direct and regulate protein synthesis in addition to coding the sequence that is translated into protein. The regulatory sequence that is before (upstream (-), towards the 5' DNA end) the coding sequence is called 5' untranslated region (5'UTR), and sequence found following (downstream (+), towards the 3' DNA end) the coding sequence is called 3' untranslated region (3'UTR). Transcription has some proofreading mechanisms, but they are fewer and less effective than the controls for copying DNA; therefore, transcription has a lower copying fidelity than DNA replication.[1]
As in DNA replication, RNA is synthesized in the 5' → 3' direction (from the point of view of the growing RNA transcript). Only one of the two DNA strands is transcribed. This strand is called the template strand, because it provides the template for ordering the sequence of nucleotides in an RNA transcript. The other strand is called the coding strand, because its sequence is the same as the newly created RNA transcript (except for uracil being substituted for thymine). The DNA template strand is read 3' → 5' by RNA polymerase and the new RNA strand is synthesized in the 5'→ 3' direction.
A polymerase binds to the 3' end of a gene (promoter) on the DNA template strand and travels toward the 5' end.
Transcription is divided into 5 stages: pre-initiation, initiation, promoter clearance, elongation and termination.
Prokaryotic vs. eukaryotic transcription
- Prokaryotic transcription occurs in the cytoplasm alongside translation. Eukaryotic transcription is localized to the nucleus, where it is separated from the cytoplasm by the nuclear membrane. The transcript is then transported into the cytoplasm where translation occurs.
- Another important difference is that eukaryotic DNA is wound around histones to form nucleosomes and packaged as chromatin. Chromatin has a strong influence on the accessibility of the DNA to transcription factors and the transcriptional machinery including RNA polymerase.
- In prokaryotes, mRNA is not usually modified. Eukaryotic mRNA is modified through RNA splicing, 5' end capping (5' cap), and the addition of a polyA tail.[2]
Major steps
Pre-initiation
In eukaryotes, RNA polymerase, and therefore the initiation of transcription, requires the presence of a core promoter sequence in the DNA. Promoters are regions of DNA which promote transcription and are found around -10 to -35 bp upstream from the start site of transcription. Core promoters are sequences within the promoter which are essential for transcription initiation. RNA polymerase is able to bind to core promoters in the presence of various specific transcription factors.
The most common type of core promoter in eukaryotes is a short DNA sequence known as a TATA box. The TATA box, as a core promoter, is the binding site for a transcription factor known as TATA binding protein (TBP), which is itself a subunit of another transcription factor, called Transcription Factor II D (TFIID). After TFIID binds to the TATA box via the TBP, five more transcription factors and RNA polymerase combine around the TATA box in a series of stages to form what is known as the preinitiation complex. One such transcription factor has helicase activity and so is involved in the separating of opposing strands of double-stranded DNA to provide access to a single-stranded DNA template. However only a low, or basal, rate of transcription is driven by this preintiation complex. Other proteins known as activators and repressors, along with any associated coactivators or corepressors, may further enhance or inhibit transcription.
Archaea transcription preinitiation is essentially homologous to that of eukaryotes, but much less complex.[3] The archaeal preinitiation complex also tend to assemble at a TATA-box binding site, however in archaea this complex is made up of only RNA polymerase II, TBP, and TFB, the archaeal homologue of eukaryotic transcription factor II B (TFIIB).[4][5]
Initiation

In bacteria, transcription begins with the binding of RNA polymerase to the promoter in DNA. The RNA polymerase is a core enzyme consisting of five subunits: 2 α subunits, 1 β subunit, 1 β' subunit, and 1 ω subunit. At the start of initiation, the core enzyme is associated with a sigma factor (number 70) that aids in finding the appropriate -35 and -10 basepairs downstream of promoter sequences.
Transcription initiation is far more complex in eukaryotes, the main difference being that eukaryotic polymerases do not directly recognize their core promoter sequences. In eukaryotes, a collection of proteins called transcription factors mediate the binding of RNA polymerase and the initiation of transcription. Only after certain transcription factors are attached to the promoter does the RNA polymerase bind to it. The completed assembly of transcription factors and RNA polymerase bind to the promoter, called transcription initiation complex. Transcription in archaea is similar to transcription in eukaryotes.[6]
Promoter clearance
After the first bond is synthesized the RNA polymerase must clear the promoter. During this time there is a tendency to release the RNA transcript and produce truncated transcripts. This is called abortive initiation and is common for both eukaryotes[7] and prokaroytes[8]. Once the transcript reaches approximately 23 nucleotides it no longer slips and elongation can occur. This is an ATP dependent process.
Promoter clearance coincides with phosphorylation of serine 5 on the carboxy terminal domain of RNA Pol in prokaryotes, which is phosphorylated by TFIIH.
Elongation

One strand of DNA, the template strand (or noncoding strand), is used as a template for RNA synthesis. As transcription proceeds, RNA polymerase traverses the template strand and uses base pairing complementarity with the DNA template to create an RNA copy. Although RNA polymerase traverses the template strand from 3' → 5', the coding (non-template) strand is usually used as the reference point, so transcription is said to go from 5' → 3'. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that thymines are replaced with uracils, and the nucleotides are composed of a ribose (5-carbon) sugar where DNA has deoxyribose (one less oxygen atom) in its sugar-phosphate backbone).
Unlike DNA replication, mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be produced from a single copy of a gene. This step also involves a proofreading mechanism that can replace incorrectly incorporated bases.
Prokaryotic elongation starts with the "abortive initiation cycle". During this cycle RNA Polymerase will synthesize mRNA fragments 2-12 nucleotides long. This continues to occur until the σ factor rearranges, which results in the transcription elongation complex (which gives a 35 bp moving footprint). The σ factor is released before 80 nucleotides of mRNA are synthesized.
In Eukaryotic transcription the polymerase can experience pauses. These pauses may be intrinsic to the RNA polymerase or due to chromatin structure. Often the polymerase pauses to allow appropriate RNA editing factors to bind.
Termination

Bacteria use two different strategies for transcription termination: in Rho-independent transcription termination, RNA transcription stops when the newly synthesized RNA molecule forms a G-C rich hairpin loop, followed by a run of U's, which makes it detach from the DNA template. In the "Rho-dependent" type of termination, a protein factor called "Rho" destabilizes the interaction between the template and the mRNA, thus releasing the newly synthesized mRNA from the elongation complex. Transcription termination in eukaryotes is less well understood. It involves cleavage of the new transcript, followed by template-independent addition of As at its new 3' end, in a process called polyadenylation.
Measuring and detecting transcription
Transcription can be measured and detected in a variety of ways:
- Nuclear Run-on assay, measures the relative abundance of newly formed transcripts
- RNase protection assay and ChIP-Chip of RNAP, detect active transcription sites
- RT-PCR, measures the absolute abundance of total or nuclear RNA levels, which may however differ from transcription rates
- DNA microarrays measures the relative abundance of the global total or nuclear RNA levels, which may however differ from transcription rates
- In situ hybridization, detects the presence of a transcript.
- MS2 tagging, by incorporating RNA stem loops, such as MS2, into a gene, these become incorporated into newly synthesized RNA. The stem loops can then be detected using a fusion of GFP and the MS2 coat protein, which has a high affinity, sequence specific interaction with the MS2 stem loops. The recruitment of GFP to the site of transcription is visualised as a single fluorescent spot. This remarkable new approach has revealed that transcription occurs in discontinuous bursts, or pulses (see Transcriptional bursting). With the notable exception of in situ techniques, most other methods provide cell population averages, and are not capable of detecting this fundamental property of genes[9].
- Northern blot The traditional method, and until the advent of RNA-Seq, probably the most quantitative
- RNA-Seq applies next-generation sequencing techniques to sequence whole transcriptomes, which allows the measurement of relative abundance of RNA, as well as the detection of additional variations such as fusion genes, post-translational edits and novel splice sites
Transcription factories
Active transcription units are clustered in the nucleus, in discrete sites called ‘transcription factories’. Such sites could be visualized after allowing engaged polymerases to extend their transcripts in tagged precursors (Br-UTP or Br-U), and immuno-labeling the tagged nascent RNA. Transcription factories can also be localized using fluorescence in situ hybridization, or marked by antibodies directed against polymerases. There are ~10,000 factories in the nucleoplasm of a HeLa cell, among which are ~8,000 polymerase II factories and ~2,000 polymerase III factories. Each polymerase II factor contains ~8 polymerases. As most active transcription units are associated with only one polymerase, each factory will be associated with ~8 different transcription units. These units might be associated through promoters and/or enhancers, with loops forming a ‘cloud’ around the factor.
History
A molecule which allows the genetic material to be realized as a protein was first hypothesized by Jacob and Monod. RNA synthesis by RNA polymerase was established in vitro by several laboratories by 1965; however, the RNA synthesized by these enzymes had properties that suggested the existence of an additional factor needed to terminate transcription correctly.
In 1972, Walter Fiers became the first person to actually prove the existence of the terminating enzyme.
Roger D. Kornberg won the 2006 Nobel Prize in Chemistry "for his studies of the molecular basis of eukaryotic transcription".[10]
Reverse transcription
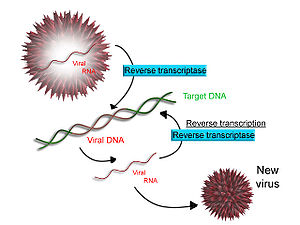
Some viruses (such as HIV, the cause of AIDS), have the ability to transcribe RNA into DNA. HIV has an RNA genome that is duplicated into DNA. The resulting DNA can be merged with the DNA genome of the host cell. The main enzyme responsible for synthesis of DNA from an RNA template is called reverse transcriptase. In the case of HIV, reverse transcriptase is responsible for synthesizing a complementary DNA strand (cDNA) to the viral RNA genome. An associated enzyme, ribonuclease H, digests the RNA strand, and reverse transcriptase synthesises a complementary strand of DNA to form a double helix DNA structure. This cDNA is integrated into the host cell's genome via another enzyme (integrase) causing the host cell to generate viral proteins which reassemble into new viral particles. Subsequently, the host cell undergoes programmed cell death (apoptosis).
Some eukaryotic cells contain an enzyme with reverse transcription activity called telomerase. Telomerase is a reverse transcriptase that lengthens the ends of linear chromosomes. Telomerase carries an RNA template from which it synthesizes DNA repeating sequence, or "junk" DNA. This repeated sequence of "junk" DNA is important because every time a linear chromosome is duplicated, it is shortened in length. With "junk" DNA at the ends of chromosomes, the shortening eliminates some repeated, or junk sequence, rather than the protein-encoding DNA sequence that is further away from the chromosome ends. Telomerase is often activated in cancer cells to enable cancer cells to duplicate their genomes without losing important protein-coding DNA sequence. Activation of telomerase could be part of the process that allows cancer cells to become technically immortal.
References
- ^ Berg J, Tymoczko JL, Stryer L (2006). Biochemistry (6th ed.). San Francisco: W. H. Freeman. ISBN 0716787245.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Robert J. Brooker Genetics: analysis and principles. 2nd edition. (New York: McGraw-Hill 2005) Chapter 12 "Gene transcription and RNA modification" pp. 318-325.
- ^ Littlefield, O., Korkhin, Y., and Sigler, P.B. (1999). "The structural basis for the oriented assembly of a TBP/TFB/promoter complex". PNAS. 96: 13668–13673. doi:10.1073/pnas.96.24.13668.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hausner, W; Thomm, M (2001). "Events during Initiation of Archaeal Transcription: Open Complex Formation and DNA-Protein Interactions". Journal of Bacteriology. 183 (10): 3025–3031. doi:10.1128/JB.183.10.3025-3031.2001. PMID 11325929.
{{cite journal}}: More than one of|author=and|last1=specified (help) - ^ Qureshi, SA; Bell, SD; Jackson, SP (1997). "Factor requirements for transcription in the archaeon Sulfolobus shibatae". EMBO Journal. 16 (10): 2927–2936. doi:10.1093/emboj/16.10.2927. PMID 9184236.
{{cite journal}}: More than one of|author=and|last1=specified (help) - ^ Mohamed Ouhammouch, Robert E. Dewhurst, Winfried Hausner, Michael Thomm, and E. Peter Geiduschek (2003). "Activation of archaeal transcription by recruitment of the TATA-binding protein". Proceedings of the National Academy of Sciences of the United States of America. 100 (9): 5097–5102. doi:10.1073/pnas.0837150100. PMC 154304. PMID 12692306.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 12213653, please use {{cite journal}} with
|pmid=12213653instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1126/science.1169237, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1126/science.1169237instead. - ^ Raj, A. and van Oudenaarden, A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216-26.
- ^ "Chemistry 2006". Nobel Foundation. Retrieved 2007-03-29.
See also
- Genetics
- Molecular biology
- Translation - process of decoding RNA to form polypeptides.
- Splicing - process of removing introns from precursor messenger RNA (pre-mRNA) to make form messenger RNA (mRNA).
- Reverse transcription - process viruses use to make DNA from RNA
- Crick's central dogma - DNA is transcribed to RNA which is translated to polypeptides, never the other way around.
Further reading
- Lehninger Principles of Biochemistry, 5th edition, David L. Nelson & Michael M. Cox
- Principles of Nuclear Structure and Function, Peter R. Cook
- Essential Genetics, Peter J. Russell
External links
- Interactive Java simulation of transcription initiation. From Center for Models of Life at the Niels Bohr Institute.
- Interactive Java simulation of transcription interference--a game of promoter dominance in bacterial virus. From Center for Models of Life at the Niels Bohr Institute.
- Comparison of prokaryotic and eukaryotic transcription
