Cancer epigenetics: Difference between revisions
Added material indicating role of epigenetic changes in DNA repair |
|||
| Line 37: | Line 37: | ||
===miRNA, DNA repair and cancer=== |
===miRNA, DNA repair and cancer=== |
||
DNA damage is considered to be the primary underlying cause of cancer.<ref name=BernsteinPrasad>Bernstein C, Prasad AR, Nfonsam V, Bernstein H. (2013). DNA Damage, DNA Repair and Cancer, New Research Directions in DNA Repair, Prof. Clark Chen (Ed.), ISBN: 978-953-51-1114-6, InTech, http://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-damage-dna-repair-and-cancer</ref> If DNA repair is deficient, damage tends to accumulate in DNA. Such DNA damage can cause [[mutational]] errors during [[DNA replication]] due to error-prone [[DNA repair#Translesion sythesis|translesion synthesis]]. Accumulated DNA damage can also cause [[Epigenetics|epigenetic]] alterations due to errors during DNA repair.<ref name= |
DNA damage is considered to be the primary underlying cause of cancer.<ref name=BernsteinPrasad>Bernstein C, Prasad AR, Nfonsam V, Bernstein H. (2013). DNA Damage, DNA Repair and Cancer, New Research Directions in DNA Repair, Prof. Clark Chen (Ed.), ISBN: 978-953-51-1114-6, InTech, http://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-damage-dna-repair-and-cancer</ref> If DNA repair is deficient, damage tends to accumulate in DNA. Such DNA damage can cause [[mutational]] errors during [[DNA replication]] due to error-prone [[DNA repair#Translesion sythesis|translesion synthesis]]. Accumulated DNA damage can also cause [[Epigenetics|epigenetic]] alterations due to errors during DNA repair.<ref name=Hagan>O'Hagan HM, Mohammad HP, Baylin SB. (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island" ''PLoS Genet'' 4(8) e1000155. {{DOI|10.1371/journal.pgen.1000155}} PMID 18704159</ref><ref name=Cuozzo>Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV (2007). DNA damage, homology-directed repair, and DNA methylation" ''PLoS Genet'' 3(7) e110. {{DOI|10.1371/journal.pgen.0030110}} PMID 17616978</ref> Such mutations and epigenetic alterations can give rise to [[cancer]] (see [[Neoplasm#Malignant neoplasms|malignant neoplasms]]). |
||
[[Germ line]] mutations in DNA repair genes cause only 2–5% of [[colon cancer]] cases.<ref>Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010). Hereditary and familial colon cancer" ''Gastroenterology'' 138(6) 2044-2058. doi: 10.1053/j.gastro.2010.01.054. PMID 20420945</ref> However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important [[causality|causal]] factor for these cancers. |
[[Germ line]] mutations in DNA repair genes cause only 2–5% of [[colon cancer]] cases.<ref>Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010). Hereditary and familial colon cancer" ''Gastroenterology'' 138(6) 2044-2058. doi: 10.1053/j.gastro.2010.01.054. PMID 20420945</ref> However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important [[causality|causal]] factor for these cancers. |
||
| Line 48: | Line 48: | ||
In contrast to the previous example, where under-expression of miRNAs indirectly caused reduced expression of DNA repair genes, in some cases over-expression of certain miRNAs may directly reduce expression of specific DNA repair proteins. Wan et al.<ref>{{cite journal |author=Wan G, Mathur R, Hu X, Zhang X, Lu X |title=miRNA response to DNA damage |journal=Trends Biochem. Sci. |volume=36 |issue=9 |pages=478–84 |date=September 2011 |pmid=21741842 |pmc=3532742 |doi=10.1016/j.tibs.2011.06.002 |url= http://www.ncbi.nlm.nih.gov/pubmed/21741842}}</ref> referred to 6 DNA repair genes that are directly targeted by the miRNAs indicated: ''ATM'' (miR-421), ''RAD52'' (miR-210, miR-373), ''RAD23B'' (miR-373), ''MSH2'' (miR-21), ''BRCA1'' (miR-182) and ''P53'' (miR-504, miR-125b). More recently, Tessitore et al.<ref name="pmid24616890">{{cite journal |author=Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E |title=MicroRNAs in the DNA Damage/Repair Network and Cancer |journal=Int J Genomics |volume=2014 |issue= |pages=820248 |year=2014 |pmid=24616890 |pmc=3926391 |doi=10.1155/2014/820248 |url=http://dx.doi.org/10.1155/2014/820248}}</ref> listed further DNA repair genes that are directly targeted by additional miRNAs, including ''ATM'' (miR-18a, miR-101), ''DNA-PK'' (miR-101), ''ATR'' (miR-185), ''Wip1'' (miR-16), ''MLH1, MSH2'' and ''MSH6'' (miR-155), ERCC3 and ERCC4 (miR-192) and ''UNG2'' (mir-16, miR-34c and miR-199a). Of these miRNAs, miR-16, miR-18a, miR-21, miR-34c, miR-125b, miR-101, miR-155, miR-182, miR-185 and miR-192 are among those identified by Schnekenburger and Diederich<ref name=Schnekenburger /> as over-expressed in colon cancer through epigenetic hypomethylation. Over expression of any one of these miRNAs can cause reduced expression of its target DNA repair gene. |
In contrast to the previous example, where under-expression of miRNAs indirectly caused reduced expression of DNA repair genes, in some cases over-expression of certain miRNAs may directly reduce expression of specific DNA repair proteins. Wan et al.<ref>{{cite journal |author=Wan G, Mathur R, Hu X, Zhang X, Lu X |title=miRNA response to DNA damage |journal=Trends Biochem. Sci. |volume=36 |issue=9 |pages=478–84 |date=September 2011 |pmid=21741842 |pmc=3532742 |doi=10.1016/j.tibs.2011.06.002 |url= http://www.ncbi.nlm.nih.gov/pubmed/21741842}}</ref> referred to 6 DNA repair genes that are directly targeted by the miRNAs indicated: ''ATM'' (miR-421), ''RAD52'' (miR-210, miR-373), ''RAD23B'' (miR-373), ''MSH2'' (miR-21), ''BRCA1'' (miR-182) and ''P53'' (miR-504, miR-125b). More recently, Tessitore et al.<ref name="pmid24616890">{{cite journal |author=Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E |title=MicroRNAs in the DNA Damage/Repair Network and Cancer |journal=Int J Genomics |volume=2014 |issue= |pages=820248 |year=2014 |pmid=24616890 |pmc=3926391 |doi=10.1155/2014/820248 |url=http://dx.doi.org/10.1155/2014/820248}}</ref> listed further DNA repair genes that are directly targeted by additional miRNAs, including ''ATM'' (miR-18a, miR-101), ''DNA-PK'' (miR-101), ''ATR'' (miR-185), ''Wip1'' (miR-16), ''MLH1, MSH2'' and ''MSH6'' (miR-155), ERCC3 and ERCC4 (miR-192) and ''UNG2'' (mir-16, miR-34c and miR-199a). Of these miRNAs, miR-16, miR-18a, miR-21, miR-34c, miR-125b, miR-101, miR-155, miR-182, miR-185 and miR-192 are among those identified by Schnekenburger and Diederich<ref name=Schnekenburger /> as over-expressed in colon cancer through epigenetic hypomethylation. Over expression of any one of these miRNAs can cause reduced expression of its target DNA repair gene. |
||
==Epigenetic DNA repair deficiencies in cancer== |
|||
Cancers have high levels of [[genome instability]], associated with a high frequency of [[mutation]]s. A high frequency of genomic mutations increases the likelihood of particular mutations occurring that activate oncogenes and inactivate tumor suppressor genes, leading to [[carcinogenesis]]. |
|||
On the basis of [[whole genome sequencing]], cancers are found to have thousands to hundreds of thousands of mutations in their whole genomes.<ref name="pmid23178448">{{cite journal |author=Tuna M, Amos CI |title=Genomic sequencing in cancer |journal=Cancer Lett. |volume=340 |issue=2 |pages=161–70 |date=November 2013 |pmid=23178448 |doi=10.1016/j.canlet.2012.11.004 |url=http://linkinghub.elsevier.com/retrieve/pii/S0304-3835(12)00651-9}}</ref> (Also see [[Whole genome sequencing#Mutation_frequencies_in_cancers|Mutation frequencies in cancers]].) By comparison, the mutation frequency in the whole genome between generations for humans (parent to child) is about 70 new mutations per generation.<ref>{{cite journal |author=Roach JC, Glusman G, Smit AF, ''et al.'' |title=Analysis of genetic inheritance in a family quartet by whole-genome sequencing |journal=Science |volume=328 |issue=5978 |pages=636–9 |date=April 2010 |pmid=20220176 |pmc=3037280 |doi=10.1126/science.1186802 |url=}}</ref><ref>{{cite journal |author=Campbell CD, Chong JX, Malig M, ''et al.'' |title=Estimating the human mutation rate using autozygosity in a founder population |journal=Nat. Genet. |volume=44 |issue=11 |pages=1277–81 |date=November 2012 |pmid=23001126 |pmc=3483378 |doi=10.1038/ng.2418 |url=}}</ref> |
|||
Deficiencies in DNA repair genes cause increased mutation rates. Mutation rates are strongly increased in cells defective in [[DNA mismatch repair]]<ref name=Narayanan>{{cite journal | author = Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM | title = Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2 | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 94 | issue = 7 | pages = 3122–7 |date=April 1997 | pmid = 9096356 | pmc = 20332 | doi = 10.1073/pnas.94.7.3122 }}</ref><ref name=Hegan>{{cite journal | author = Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM | title = Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6 | journal = Carcinogenesis | volume = 27 | issue = 12 | pages = 2402–8 |date=December 2006 | pmid = 16728433 | pmc = 2612936 | doi = 10.1093/carcin/bgl079 }}</ref> or in [[homologous recombination]]al repair (HRR).<ref name=Tutt>{{cite journal | author = Tutt AN, van Oostrom CT, Ross GM, van Steeg H, Ashworth A | title = Disruption of Brca2 increases the spontaneous mutation rate in vivo: synergism with ionizing radiation | journal = EMBO Rep. | volume = 3 | issue = 3 | pages = 255–60 |date=March 2002 | pmid = 11850397 | pmc = 1084010 | doi = 10.1093/embo-reports/kvf037 }}</ref> Individuals with inherited mutations in any of 34 DNA repair genes are at increased risk of cancer (see [[DNA repair-deficiency disorder#DNA_repair_defects_and_increased_cancer_risk|DNA repair defects and increased cancer risk]].) . However, such germ line mutations (which cause highly penetrant cancer syndromes) are the cause of only about 1 percent of cancers.<ref>{{Cite journal|pages=1043–1050|pmid=9353177|year=1997|last1=Fearon|first1=ER|title=Human cancer syndromes: Clues to the origin and nature of cancer|volume=278|issue=5340|journal=Science|doi=10.1126/science.278.5340.1043}}</ref> |
|||
In sporadic cancers, a deficiency in DNA repair is occasionally found to be due to a mutation in a DNA repair gene, but much more frequently reduced or absent expression of DNA repair genes is due to epigenetic alterations that reduce or silence gene expression. For example, for 113 colorectal cancers examined in sequence, only four had a [[missense mutation]] in the DNA repair gene [[O-6-methylguanine-DNA methyltransferase|''MGMT'']], while the majority had reduced ''MGMT'' expression due to methylation of the ''MGMT'' promoter region (an epigenetic alteration).<ref>{{Cite journal|pages=797–802|doi=10.1136/gut.2004.059535|pmid=15888787|title=O6-methylguanine methyltransferase in colorectal cancers: Detection of mutations, loss of expression, and weak association with G:C>A:T transitions|year=2005|last1=Halford|first1=S|journal=Gut|volume=54|issue=6|last2=Rowan|first2=A|last3=Sawyer|first3=E|last4=Talbot|first4=I|last5=Tomlinson|first5=I|pmc=1774551}}</ref> |
|||
Similarly, out of 119 cases of mismatch repair-deficient colorectal cancers that lacked DNA repair gene ''PMS2'' expression, PMS2 protein was deficient in 6 due to mutations in the ''PMS2'' gene, while in 103 cases PMS2 expression was deficient because its pairing partner MLH1 was repressed due to promoter methylation (PMS2 protein is unstable in the absence of MLH1).<ref>{{Cite journal|pages=1160–1171|doi=10.1053/j.gastro.2005.01.056|pmid=15887099|title=Immunohistochemical Analysis Reveals High Frequency of PMS2 Defects in Colorectal Cancer|year=2005|last1=Truninger|first1=Kaspar|last2=Menigatti|first2=Mirco|last3=Luz|first3=Judith|last4=Russell|first4=Anna|last5=Haider|first5=Ritva|last6=Gebbers|first6=Jan-Olaf|last7=Bannwart|first7=Fridolin|last8=Yurtsever|first8=Hueseyin|last9=Neuweiler|first9=Joerg|last10=Riehle|first10=Hans-Martin|last11=Cattaruzza|first11=Maria Sofia|last12=Heinimann|first12=Karl|last13=Schär|first13=Primo|last14=Jiricny|first14=Josef|last15=Marra|first15=Giancarlo|journal=Gastroenterology|volume=128|issue=5}}</ref> In the other 10 cases, loss of PMS2 expression was likely due to epigenetic overexpression of the microRNA, miR-155, which down-regulates MLH1.<ref>{{Cite journal|pages=6982–6987|doi=10.1073/pnas.1002472107|pmid= 20351277|title=Modulation of mismatch repair and genomic stability by miR-155|year=2010|last1=Valeri|first1=N.|last2=Gasparini|first2=P.|last3=Fabbri|first3=M.|last4=Braconi|first4=C.|last5=Veronese|first5=A.|last6=Lovat|first6=F.|last7=Adair|first7=B.|last8=Vannini|first8=I.|last9=Fanini|first9=F.|last10=Bottoni|first10=A.|last11=Costinean|first11=S.|last12=Sandhu|first12=S. K.|last13=Nuovo|first13=G. J.|last14=Alder|first14=H.|last15=Gafa|first15=R.|last16=Calore|first16=F.|last17=Ferracin|first17=M.|last18=Lanza|first18=G.|last19=Volinia|first19=S.|last20=Negrini|first20=M.|last21=McIlhatton|first21=M. A.|last22=Amadori|first22=D.|last23=Fishel|first23=R.|last24=Croce|first24=C. M.|journal=Proceedings of the National Academy of Sciences|volume=107|issue=15|pmc=2872463}}</ref> |
|||
Epigenetic defects in DNA repair genes are frequent in cancers. In the Table, multiple cancers were evaluated for reduced or absent expression of the DNA repair gene of interest, and the frequency shown is the frequency with which the cancers had an epigenetic deficiency of gene expression. Such epigenetic deficiencies are likely early-arising defects in the path to [[carcinogenesis]], since they are also frequently found (though at somewhat lower frequency) in the [[Neoplasm#Field_defects|field defect]] surrounding the cancer from which the cancer likely arose (see Table). |
|||
{| class="wikitable sortable" |
|||
|+ Frequency of epigenetic changes in DNA repair genes in sporadic cancers and in adjacent field defects |
|||
! Cancer !!Gene !!Frequency in Cancer !!Frequency in Field Defect!!Ref. |
|||
|- |
|||
!Colorectal |
|||
|MGMT || 46%||34%||<ref name="pmid16174854">{{cite journal |author=Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JP |title=MGMT promoter methylation and field defect in sporadic colorectal cancer |journal=J. Natl. Cancer Inst. |volume=97 |issue=18 |pages=1330–8 |year=2005 |month=September |pmid=16174854 |doi=10.1093/jnci/dji275 |url=http://jnci.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=16174854}}</ref> |
|||
|- |
|||
!Colorectal |
|||
|MGMT || 47%||11%||<ref name="Lee KH 2011"/> |
|||
|- |
|||
!Colorectal |
|||
|MGMT || 70%||60%||<ref name="Svrcek et al 2010">{{cite journal | author = Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, Seruca R, Gaub MP, Legrain M, Collura A, Lascols O, Tiret E, Fléjou JF, Duval A | title = Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers | journal = Gut | volume = 59 | issue = 11 | pages = 1516–26 |date=November 2010 | pmid = 20947886 | doi = 10.1136/gut.2009.194787 }}</ref> |
|||
|- |
|||
!Colorectal |
|||
|MSH2 || 13%||5%|| <ref name="pmid21706233">{{cite journal |author=Lee KH, Lee JS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Lee JH |title=Promoter methylation status of hMLH1, hMSH2, and MGMT genes in colorectal cancer associated with adenoma-carcinoma sequence |journal=Langenbecks Arch Surg |volume=396 |issue=7 |pages=1017–26 |year=2011 |month=October |pmid=21706233 |doi=10.1007/s00423-011-0812-9 |url=http://dx.doi.org/10.1007/s00423-011-0812-9}}</ref> |
|||
|- |
|||
!Colorectal |
|||
|ERCC1 || 100%||40%||<ref name=Facista /> |
|||
|- |
|||
!Colorectal |
|||
|PMS2 || 88%||50%||<ref name=Facista /> |
|||
|- |
|||
!Colorectal |
|||
|XPF || 55%||40%||<ref name=Facista /> |
|||
|- |
|||
!Head and Neck |
|||
|MGMT || 54%||38%||<ref name="Jaroslaw et al 2011">{{cite journal | author = Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer-Dubowska W | title = Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa | journal = Oral Oncol. | volume = 47 | issue = 2 | pages = 104–7 |date=February 2011 | pmid = 21147548 | doi = 10.1016/j.oraloncology.2010.11.006 }}</ref> |
|||
|- |
|||
!Head and Neck |
|||
|MLH1 || 33%||25%||<ref name="Chunlai et al 2009">{{cite journal | author = Zuo C, Zhang H, Spencer HJ, Vural E, Suen JY, Schichman SA, Smoller BR, Kokoska MS, Fan CY | title = Increased microsatellite instability and epigenetic inactivation of the hMLH1 gene in head and neck squamous cell carcinoma | journal = Otolaryngol Head Neck Surg | volume = 141 | issue = 4 | pages = 484–90 |date=October 2009 | pmid = 19786217 | doi = 10.1016/j.otohns.2009.07.007 }}</ref> |
|||
|- |
|||
!Head and Neck |
|||
|MLH1 || 31%||20%||<ref name="Tawfik et al 2011">{{cite journal | author = Tawfik HM, El-Maqsoud NM, Hak BH, El-Sherbiny YM | title = Head and neck squamous cell carcinoma: mismatch repair immunohistochemistry and promoter hypermethylation of hMLH1 gene | journal = Am J Otolaryngol | volume = 32 | issue = 6 | pages = 528–36 | year = 2011 | pmid = 21353335 | doi = 10.1016/j.amjoto.2010.11.005 }}</ref> |
|||
|- |
|||
!Stomach |
|||
|MGMT || 88%||78%||<ref name="Zou et al 2009">{{cite journal | author = Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ | title = Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions | journal = Hum. Pathol. | volume = 40 | issue = 11 | pages = 1534–42 |date=November 2009 | pmid = 19695681 | doi = 10.1016/j.humpath.2009.01.029 }}</ref> |
|||
|- |
|||
!Stomach |
|||
|MLH1 || 73%||20%||<ref name="pmid23098428">{{cite journal | author = Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G, Wani R, Wani K | title = Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley | journal = Asian Pac. J. Cancer Prev. | volume = 13 | issue = 8 | pages = 4177–81 | year = 2012 | pmid = 23098428 | doi = 10.7314/APJCP.2012.13.8.4177 }}</ref> |
|||
|- |
|||
!Esophagus |
|||
|MLH1 || 77%-100%||23%-79%||<ref name="Agarwal et al 2012">{{cite journal | author = Agarwal A, Polineni R, Hussein Z, Vigoda I, Bhagat TD, Bhattacharyya S, Maitra A, Verma A | title = Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma | journal = Int J Clin Exp Pathol | volume = 5 | issue = 5 | pages = 382–96 | year = 2012 | pmid = 22808291 | pmc = 3396065 | doi = }}</ref> |
|||
|} |
|||
===Epigenetic defects in DNA repair pathways=== |
|||
[[File:DNA damage, repair, epigenetic alteration of repair in cancer.jpg|thumb|600px|A chart of common DNA damaging agents, the lesions they cause in DNA, the repair pathways utilized to repair these lesions, many of the genes in those pathways, indication of which genes are regulated epigenetically, and which of those epigenetically regulated genes are found with reduced expression in various cancers.]] |
|||
Epigenetic control of expression has been shown for at least 26 DNA repair genes, shown in the chart in red. The chart in this section shows some frequent DNA damaging agents, examples of DNA lesions they cause, and the pathways that deal with these DNA damages. At least 169 enzymes are either directly employed in DNA repair or influence DNA repair processes.<ref>http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html</ref> Of these, 83 are directly employed in the 5 types of DNA repair processes illustrated in the chart. Forty seven of the most well studied genes central to these repair processes are also shown in the chart. As indicated by the DNA repair genes shown in red, many of the genes in these repair pathways are regulated by epigenetic mechanisms, and the expressions of these genes are frequently reduced or silent in various cancers (marked by an asterisk). Two review articles.<ref name=BernsteinPrasad /><ref>Xinrong Chen and Tao Chen (2011). Roles of MicroRNA in DNA Damage and Repair, DNA Repair, Dr. Inna Kruman (Ed.), ISBN 978-953-307-697-3, InTech, Available from: http://www.intechopen.com/books/dnarepair/roles-of-microrna-in-dna-damage-and-repair</ref> and two broad experimental survey articles<ref>Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM. (2013). MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 19(2):230-42. doi: 10.1261/rna.034926.112. PMID 23249749 http://www.ncbi.nlm.nih.gov/pubmed/23249749</ref><ref>Chaisaingmongkol J, Popanda O, Warta R, Dyckhoff G, Herpel E, Geiselhart L, Claus R, Lasitschka F, Campos B, Oakes CC, Bermejo JL, Herold-Mende C, Plass C, Schmezer P. (2012). Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma. Oncogene 31(49):5108–16. doi: 10.1038/onc.2011.660. PMID 22286769</ref> document most of these epigenetic DNA repair deficiencies. |
|||
It appears that cancers may frequently be initiated by epigenetic reductions(s) in expression of one or more DNA repair enzymes. For instance, simultaneous epigenetic deficiency of DNA repair enzymes in two DNA repair pathways (nucleotide excision repair and mismatch repair) were found in the majority of 49 colon cancers evaluated in one report. <ref name=Facista /> Reduced DNA repair likely allows accumulation of DNA damages. Error prone [[DNA repair#Translesion_synthesis|translesion synthesis]] past some of these DNA damages may give rise to a mutation with a selective advantage. A clonal patch of with a selective advantage may grow and out-compete neighboring cells, forming a [[Neoplasm#Field defects|field defect]]. While there is no obvious [[Natural selection#Cell_and_molecular_biology|selective advantage]] for a cell with reduced DNA repair, the [[epimutation]] of the DNA repair gene may be carried along as a passenger when the cells with the selectively advantageous mutation are replicated. In the cells carrying both the epimutation of the DNA repair gene and the mutation with the selective advantage, further DNA damage will accumulate, and that could, in turn, give rise to further mutations with still greater selective advantages. Epigenetic defects in DNA repair may thus contribute to the characteristic high frequency of mutations in the genome of cancers, and cause their carcinogenic progression. |
|||
While DNA damages may give rise to mutations through error prone [[DNA repair#Translesion_synthesis|translesion synthesis]], DNA damages may also give rise to epigenetic alterations through steps in the DNA repair process.<ref name=Hagan /><ref name=Cuozzo /><ref name="pmid20550933">{{cite journal |author=Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA |title=ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks |journal=Cell |volume=141 |issue=6 |pages=970–81 |year=2010 |month=June |pmid=20550933 |pmc=2920610 |doi=10.1016/j.cell.2010.04.038 |url=http://linkinghub.elsevier.com/retrieve/pii/S0092-8674(10)00492-7}}</ref><ref name="pmid24137009">{{cite journal |author=Morano A, Angrisano T, Russo G, Landi R, Pezone A, Bartollino S, Zuchegna C, Babbio F, Bonapace IM, Allen B, Muller MT, Chiariotti L, Gottesman ME, Porcellini A, Avvedimento EV |title=Targeted DNA methylation by homology-directed repair in mammalian cells. Transcription reshapes methylation on the repaired gene |journal=Nucleic Acids Res. |volume=42 |issue=2 |pages=804–21 |year=2014 |month=January |pmid=24137009 |pmc=3902918 |doi=10.1093/nar/gkt920 |url=http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=24137009}}</ref> The DNA damages that accumulate due to epigenetic DNA repair defects can be a source of increased epigenetic alterations found in many genes in cancers, and some of these epigenetic alterations may contribute to cancer progression. |
|||
==Cancer subtype specific epigenetic modifications== |
==Cancer subtype specific epigenetic modifications== |
||
Revision as of 03:44, 29 April 2014
Cancer epigenetics is the study of epigenetic modifications to the genome of cancer cells that do not involve a change in the nucleotide sequence. Epigenetic alterations are as important as genetic mutations in a cell’s transformation to cancer.[1] Mechanisms of epigenetic silencing of tumor suppressor genes and activation of oncogenes include: alteration in CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. Understanding epigenetic mechanisms holds great promise for cancer prevention, detection, and therapy.


DNA methylation
DNA methylation is probably the most well researched epigenetic mark that differs between normal cells and tumor cells in humans. The "normal" CpG methylation profile is often inverted in cells that become tumorigenic.[2] In normal cells, CpG Islands preceding gene promoters are generally unmethylated, while other individual CpG dinucleotides throughout the genome tend to be methylated. However, in cancer cells, CpG islands preceding tumor suppressor gene promoters are often hypermethylated, while CpG methylation of oncogene promoter regions and parasitic repeat sequences is often decreased.
Hypermethylation of gene promoters can result in silencing of those genes. This type of epigenetic mutation is dangerous when genes that regulate the cell cycle are silenced, allowing cells to grow and reproduce uncontrollably, leading to tumorigenesis.[2] Genes commonly found to be transcriptionally silenced due to promoter hypermethylation include: Cyclin-dependent kinase inhibitor p16, a cell-cycle inhibitor; p53, a tumor suppressor gene; MGMT, a DNA repair gene; APC, a cell cycle regulator; MLH1, a DNA-repair gene; and BRCA1, another DNA-repair gene.[2][3] Indeed, cancer cells can become addicted to the transcriptional silencing, due to promoter hypermethylation, of some key tumor suppressor genes, a process known as epigenetic addiction[4]
Hypomethylation of CpG dinucleotides in other parts of the genome leads to chromosome instability due to mechanisms such as loss of imprinting and reactivation of transposable elements.[5][6][7][8] Loss of imprinting of insulin-like growth factor gene (IGF2) increases risk of colorectal cancer and is associated with Beckwith-Wiedemann syndrome which significantly increases the risk of cancer for newborns.[9] In healthy cells, CpG dinucleotides of lower densities are found within coding and non-coding intergenic regions. Parasitic repetitive sequences, centromeres and oncogenes are often repressed through methylation.
The entire genome of a cancerous cell contains significantly less methylcytosine than the genome of a healthy cell. In fact, cancer cell genomes have 20-50% less methylation at individual CpG dinucleotides across the genome.[5][6][7][8] In cancer cells “global hypomethylation” due to disruption in DNA methyltransferases (DNMTs) may promote mitotic recombination and chromosome rearrangement, ultimately resulting in aneuploidy when the chromosomes fail to separate properly during mitosis.[5][6][7][8]
CpG island methylation is important in regulation of gene expression, yet cytosine methylation can lead directly to destabilizing genetic mutations and a precancerous cellular state. Methylated cytosines make hydrolysis of the amine group and spontaneous conversion to thymine more favorable. They can cause aberrant recruitment of chromatin proteins. Cytosine methylations change the amount of UV light absorption of the nucleotide base, creating pyrimidine dimers. When mutation results in loss of heterozygosity at tumor suppressor gene sites, these genes may become inactive. Single base pair mutations during replication can also have detrimental effects.[3]
Histone modifications
Eukaryotic DNA has a complex structure. It is generally wrapped around special proteins called histones to form a structure called a nucleosome. A nucleosome consists of 2 sets of 4 histones: H2A, H2B, H3, and H4. Additionally, histone H1 contributes to DNA packaging outside of the nucleosome. Certain histone modifying enzymes can add or remove functional groups to the histones, and these modifications influence the level of transcription of the genes wrapped around those histones and the level of DNA replication. Therefore, one might expect the histone modification profiles of healthy and cancerous cells to differ.
In comparison to healthy cells, cancerous cells have been seen to exhibit decreased monoacetylated and trimethylated forms of histone H4 (decreased H4ac and H4me3).[10] In mouse models, scientists have noticed that the loss of histone H4 acetylation and trimethylation actually increases as tumor growth continues.[10] Interestingly, loss of histone H4 Lysine 16 acetylation (H4K16ac), which is a mark of aging at the telomeres, specifically loses its acetylation. Some scientists hope this particular loss of histone acetylation might be battled with a histone deacetylase (HDAC) inhibitor specific for SIRT1, an HDAC specific for H4K16.[2][11]
Other histone marks associated with tumorigenesis include increased deacetylation (decreased acetylation) of histones H3 and H4, decreased trimethylation of histone H3 Lysine 4 (H3K4me3), and increased monomethylation of histone H3 Lysine 9 (H3K9me) and trimethylation of histone H3 Lysine 27 (H3K27me3). These histone modifications can silence tumor suppressor genes despite the drop in methylation of the gene’s CpG island (an event that normally activates genes).[12][13]
Some research has focused on blocking the action of BRD4 on acetylated histones, which has been shown to increase the expression of the Myc protein, implicated in several cancers. The development process of the drug to bind to BRD4 is noteworthy for the collaborative, open approach the team is taking.[14]
The tumor suppressor gene p53 regulates DNA repair and can induce apoptosis in dysregulated cells. E Soto-Reyes and F Recillas-Targa elucidated the importance of the CTCF protein in regulating p53 expression.[15] CTCF, or CCCTC binding factor, is a zinc finger protein that insulates the p53 promoter from accumulating repressive histone marks. In certain types of cancer cells, the CTCF protein does not bind normally, and the p53 promoter accumulates repressive histone marks, causing p53 expression to decrease.[15]
Mutations in the epigenetic machinery itself may occur as well, potentially responsible for the changing epigenetic profiles of cancerous cells. For example, the decrease in H4K16ac may be caused by either a decrease in activity of a histone acetyltransferases (HATs) or an increase in deacetylation by SIRT1.[2] Likewise, an inactivating frameshift mutation in HDAC2, a histone deacetylase that acts on many histone-tail lysines, has been associated with cancers showing altered histone acetylation patterns.[16] These findings indicate a promising mechanism for altering epigenetic profiles through enzymatic inhibition or enhancement.
DNA damage, caused by UV light, ionizing radiation, environmental toxins, and metabolic chemicals, can also lead to genomic instability and cancer. The DNA damage response to double strand DNA breaks (DSB) is mediated in part by histone modifications. At a DSB, MRE11-RAD50-NBS1 (MRN) protein complex recruits ataxia telangiectasia mutated (ATM) kinase which phosphorylates Serine 129 of Histone 2A. MDC1, mediator of DNA damage checkpoint 1, binds to the phosphopeptide, and phosphorylation of H2AX may spread by a positive feedback loop of MRN-ATM recruitment and phosphorylation. TIP60 acetylates the γH2AX, which is then polyubiquitylated. RAP80, a subunit of the DNA repair breast cancer type 1 susceptibility protein complex (BRCA1-A), binds ubiquitin attached to histones. BRCA1-A activity arrests the cell cycle at the G2/M checkpoint, allowing time for DNA repair, or apoptosis may be initiated.[17]
MicroRNA gene silencing
In mammals, microRNA (miRNA) regulates around 60% of the transcriptional activity of protein-encoding genes. Some miRNAs have also been found to undergo methylation-associated silencing in cancerous cells.[18][19] Let-7 and miR15/16 play important roles in down-regulating RAS and BCL2 oncogenes, and their silencing has been found in cancer cells.[9] A decrease in expression of miR-125b1, a miRNA that functions as a tumor suppressor, was observed in prostate, ovarian, breast and glial cell cancers. In vitro experiments have shown that miR-125b1 targets two genes, HER2/neu and ESR1, that are linked to breast cancer. DNA methylation, specifically hypermethylation, is one of the main ways that the miR-125b1 is epigenetically silenced. In patients with breast cancer, hypermethylation of CpG islands located proximal to the transcription start site was observed. Loss of CTCF binding and an increase in repressive histone marks, H3K9me3 and H3K27me3, correlates with DNA methylation and miR-125b1 silencing. Mechanistically, CTCF may function as a boundary element to stop the spread of DNA methylation. Results from experiments conducted by Soto-Reyes indicate a negative effect of methylation on the function and expression of miR-125b1, therefore Soto-Reyes and his team were able to conclude that DNA methylation has a part in silencing the gene. Furthermore, results show that some miRNA’s are epigenetically silenced early on in breast cancer, and therefore these miRNA’s could potentially be useful as tumor markers.[20]
miRNA, DNA repair and cancer
DNA damage is considered to be the primary underlying cause of cancer.[21] If DNA repair is deficient, damage tends to accumulate in DNA. Such DNA damage can cause mutational errors during DNA replication due to error-prone translesion synthesis. Accumulated DNA damage can also cause epigenetic alterations due to errors during DNA repair.[22][23] Such mutations and epigenetic alterations can give rise to cancer (see malignant neoplasms).
Germ line mutations in DNA repair genes cause only 2–5% of colon cancer cases.[24] However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important causal factor for these cancers.
Among 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1, most were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene.[25] However, up to 15% of the MLH1-deficiencies in sporadic colon cancers appeared to be due to over-expression of the microRNA miR-155, which represses MLH1 expression.[26] In 29–66%[27][28] of glioblastomas, DNA repair is deficient due to epigenetic methylation of the MGMT gene, which reduces protein expression of MGMT. However, for 28% of glioblastomas, the MGMT protein is deficient but the MGMT promoter is not methylated.[27] In the glioblastomas without methylated MGMT promoters, the level of microRNA miR-181d is inversely correlated with protein expression of MGMT and the direct target of miR-181d is the MGMT mRNA 3’UTR (the three prime untranslated region of MGMT mRNA).[27] Thus, in 28% of glioblastomas, increased expression of miR-181d and reduced expression of DNA repair enzyme MGMT may be a causal factor.
HMGA proteins (HMGA1a, HMGA1b and HMGA2) are implicated in cancer, and expression of these proteins is regulated by microRNAs. HMGA expression is almost undetectable in differentiated adult tissues but is elevated in many cancers. HGMA proteins are polypeptides of ~100 amino acid residues characterized by a modular sequence organization. These proteins have three highly positively-charged regions, termed AT hooks, that bind the minor groove of AT-rich DNA stretches in specific regions of DNA. Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinoma, show a strong increase of HMGA1a and HMGA1b proteins.[29] Transgenic mice with HMGA1 targeted to lymphoid cells develop aggressive lymphoma, showing that high HMGA1 expression is not only associated with cancers, but that the HMGA1 gene can act as an oncogene to cause cancer.[30] Baldassarre et al.,[31] showed that HMGA1 protein binds to the promoter region of DNA repair gene BRCA1 and inhibits BRCA1 promoter activity. They also showed that while only 11% of breast tumors had hypermethylation of the BRCA1 gene, 82% of aggressive breast cancers have low BRCA1 protein expression, and most of these reductions were due to chromatin remodeling by high levels of HMGA1 protein. HMGA2 protein specifically targets the promoter of ERCC1, thus reducing expression of this DNA repair gene.[32] ERCC1 protein expression was deficient in 100% of 47 evaluated colon cancers (though the extent to which HGMA2 was involved is not known).[33] Palmieri et al.[34] showed that, in normal tissues, HGMA1 and HMGA2 genes are targeted (and thus strongly reduced in expression) by miR-15, miR-16, miR-26a, miR-196a2 and Let-7a. However, each of these HMGA-targeting miRNAs are drastically reduced in almost all human pituitary adenomas studied, when compared with the normal pituitary gland. Consistent with the down-regulation of these HMGA-targeting miRNAs, an increase in the HMGA1 and HMGA2-specific mRNAs was observed. Three of these microRNAs (miR-16, miR-196a and Let-7a)[35][36] have methylated promoters and therefore low expression in colon cancer. For two of these, miR-15 and miR-16, the coding regions are epigenetically silenced in cancer due to histone deacetylase activity.[37] When these microRNAs are expressed at a low level, then HMGA1 and HMGA2 proteins are expressed at a high level. HMGA1 and HMGA2 target (reduce expression of) BRCA1 and ERCC1 DNA repair [38] genes. Thus DNA repair can be reduced, likely contributing to cancer progression.[21]
In contrast to the previous example, where under-expression of miRNAs indirectly caused reduced expression of DNA repair genes, in some cases over-expression of certain miRNAs may directly reduce expression of specific DNA repair proteins. Wan et al.[39] referred to 6 DNA repair genes that are directly targeted by the miRNAs indicated: ATM (miR-421), RAD52 (miR-210, miR-373), RAD23B (miR-373), MSH2 (miR-21), BRCA1 (miR-182) and P53 (miR-504, miR-125b). More recently, Tessitore et al.[40] listed further DNA repair genes that are directly targeted by additional miRNAs, including ATM (miR-18a, miR-101), DNA-PK (miR-101), ATR (miR-185), Wip1 (miR-16), MLH1, MSH2 and MSH6 (miR-155), ERCC3 and ERCC4 (miR-192) and UNG2 (mir-16, miR-34c and miR-199a). Of these miRNAs, miR-16, miR-18a, miR-21, miR-34c, miR-125b, miR-101, miR-155, miR-182, miR-185 and miR-192 are among those identified by Schnekenburger and Diederich[36] as over-expressed in colon cancer through epigenetic hypomethylation. Over expression of any one of these miRNAs can cause reduced expression of its target DNA repair gene.
Epigenetic DNA repair deficiencies in cancer
Cancers have high levels of genome instability, associated with a high frequency of mutations. A high frequency of genomic mutations increases the likelihood of particular mutations occurring that activate oncogenes and inactivate tumor suppressor genes, leading to carcinogenesis.
On the basis of whole genome sequencing, cancers are found to have thousands to hundreds of thousands of mutations in their whole genomes.[41] (Also see Mutation frequencies in cancers.) By comparison, the mutation frequency in the whole genome between generations for humans (parent to child) is about 70 new mutations per generation.[42][43]
Deficiencies in DNA repair genes cause increased mutation rates. Mutation rates are strongly increased in cells defective in DNA mismatch repair[44][45] or in homologous recombinational repair (HRR).[46] Individuals with inherited mutations in any of 34 DNA repair genes are at increased risk of cancer (see DNA repair defects and increased cancer risk.) . However, such germ line mutations (which cause highly penetrant cancer syndromes) are the cause of only about 1 percent of cancers.[47]
In sporadic cancers, a deficiency in DNA repair is occasionally found to be due to a mutation in a DNA repair gene, but much more frequently reduced or absent expression of DNA repair genes is due to epigenetic alterations that reduce or silence gene expression. For example, for 113 colorectal cancers examined in sequence, only four had a missense mutation in the DNA repair gene MGMT, while the majority had reduced MGMT expression due to methylation of the MGMT promoter region (an epigenetic alteration).[48] Similarly, out of 119 cases of mismatch repair-deficient colorectal cancers that lacked DNA repair gene PMS2 expression, PMS2 protein was deficient in 6 due to mutations in the PMS2 gene, while in 103 cases PMS2 expression was deficient because its pairing partner MLH1 was repressed due to promoter methylation (PMS2 protein is unstable in the absence of MLH1).[49] In the other 10 cases, loss of PMS2 expression was likely due to epigenetic overexpression of the microRNA, miR-155, which down-regulates MLH1.[50]
Epigenetic defects in DNA repair genes are frequent in cancers. In the Table, multiple cancers were evaluated for reduced or absent expression of the DNA repair gene of interest, and the frequency shown is the frequency with which the cancers had an epigenetic deficiency of gene expression. Such epigenetic deficiencies are likely early-arising defects in the path to carcinogenesis, since they are also frequently found (though at somewhat lower frequency) in the field defect surrounding the cancer from which the cancer likely arose (see Table).
| Cancer | Gene | Frequency in Cancer | Frequency in Field Defect | Ref. |
|---|---|---|---|---|
| Colorectal | MGMT | 46% | 34% | [51] |
| Colorectal | MGMT | 47% | 11% | [52] |
| Colorectal | MGMT | 70% | 60% | [53] |
| Colorectal | MSH2 | 13% | 5% | [54] |
| Colorectal | ERCC1 | 100% | 40% | [33] |
| Colorectal | PMS2 | 88% | 50% | [33] |
| Colorectal | XPF | 55% | 40% | [33] |
| Head and Neck | MGMT | 54% | 38% | [55] |
| Head and Neck | MLH1 | 33% | 25% | [56] |
| Head and Neck | MLH1 | 31% | 20% | [57] |
| Stomach | MGMT | 88% | 78% | [58] |
| Stomach | MLH1 | 73% | 20% | [59] |
| Esophagus | MLH1 | 77%-100% | 23%-79% | [60] |
Epigenetic defects in DNA repair pathways
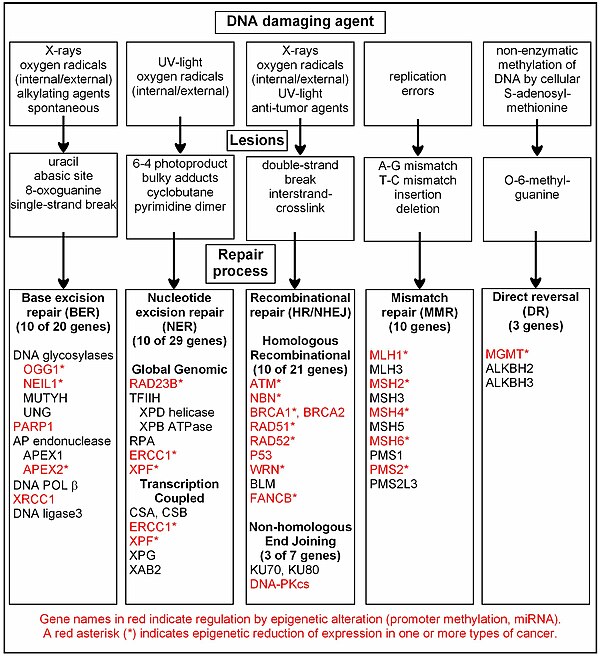
Epigenetic control of expression has been shown for at least 26 DNA repair genes, shown in the chart in red. The chart in this section shows some frequent DNA damaging agents, examples of DNA lesions they cause, and the pathways that deal with these DNA damages. At least 169 enzymes are either directly employed in DNA repair or influence DNA repair processes.[61] Of these, 83 are directly employed in the 5 types of DNA repair processes illustrated in the chart. Forty seven of the most well studied genes central to these repair processes are also shown in the chart. As indicated by the DNA repair genes shown in red, many of the genes in these repair pathways are regulated by epigenetic mechanisms, and the expressions of these genes are frequently reduced or silent in various cancers (marked by an asterisk). Two review articles.[21][62] and two broad experimental survey articles[63][64] document most of these epigenetic DNA repair deficiencies.
It appears that cancers may frequently be initiated by epigenetic reductions(s) in expression of one or more DNA repair enzymes. For instance, simultaneous epigenetic deficiency of DNA repair enzymes in two DNA repair pathways (nucleotide excision repair and mismatch repair) were found in the majority of 49 colon cancers evaluated in one report. [33] Reduced DNA repair likely allows accumulation of DNA damages. Error prone translesion synthesis past some of these DNA damages may give rise to a mutation with a selective advantage. A clonal patch of with a selective advantage may grow and out-compete neighboring cells, forming a field defect. While there is no obvious selective advantage for a cell with reduced DNA repair, the epimutation of the DNA repair gene may be carried along as a passenger when the cells with the selectively advantageous mutation are replicated. In the cells carrying both the epimutation of the DNA repair gene and the mutation with the selective advantage, further DNA damage will accumulate, and that could, in turn, give rise to further mutations with still greater selective advantages. Epigenetic defects in DNA repair may thus contribute to the characteristic high frequency of mutations in the genome of cancers, and cause their carcinogenic progression.
While DNA damages may give rise to mutations through error prone translesion synthesis, DNA damages may also give rise to epigenetic alterations through steps in the DNA repair process.[22][23][65][66] The DNA damages that accumulate due to epigenetic DNA repair defects can be a source of increased epigenetic alterations found in many genes in cancers, and some of these epigenetic alterations may contribute to cancer progression.
Cancer subtype specific epigenetic modifications
Prostate cancer
Prostate cancer kills around 35,000 men yearly, and about 220,000 men are diagnosed with prostate cancer per year, in North America alone.[67] Prostate cancer is the second leading cause of cancer-caused fatalities in men, and within a man’s lifetime, one in six men will have the disease.[67] Prostate cancer has been associated with gene silencing by CpG island hypermethylation. The GSTP1 gene has been found to defend prostate cells against genomic damage that is caused by different oxidants or carcinogens.[68] This suggests that the silencing of this gene will permit genetic damage to the prostate by oxidants and carcinogens. Methylation of cytosines within a CpG island is a causative factor in gene silencing. Modern epigenetics have correlated CpG island hypermethylation with unexpressed genes. Hypermethylation of the CpG island within the promoter region of the GSTP1 gene has been found to occur in more than 90% of prostate cancers.[68] The high percentage of occurrence demonstrates that hypermethylation of the CpG island within the promoter region of the GSTP1 gene is a highly causative factor of prostate carcinogenesis. In prostate cancer, many other genes have been found to be hypermethylated. An experiment using quantitative real-time methylation-specific polymerase chain reaction (PCR) viewed 16 different genes of prostate cancer that were hypermethylated in high frequencies of the CpG islands in GSTP1, APC, RASSF1a, PTGS2, and MDRI[clarification needed], but in normal prostate tissues almost no methylation was found.[68] Polymerase chain reaction shows the fragment sizes of DNA of interest. Hypermethylation of the genes were shown to be unexpressed. This demonstrates that the hypermethylation of the 16 different genes show no expression of these genes in prostate cancer, and that these genes are necessary for a normal functioning disease-free prostate.
Cervical cancer
The second most common malignant tumor in women is invasive cervical cancer (ICC) and more than 50% of all invasive cervical cancer (ICC) is caused by oncongenic human papillomavirus 16 (HPV16).[69] Furthermore, cervix intraepithelial neoplasia (CIN) is primarily caused by oncogenic HPV16.[69] As in many cases, the causative factor for cancer does not always take a direct route from infection to the development of cancer. Genomic methylation patterns have been associated with invasive cervical cancer. Within the HPV16L1 region, 14 tested CpG sites have significantly higher methylation in CIN3+ than in HPV16 genomes of women without CIN3.[69] Only 2/16 CpG sites tested in HPV16 upstream regulatory region were found to have association with increased methylation in CIN3+.[69] This suggests that the direct route from infection to cancer is sometimes detoured to a precancerous state in cervix intraepithelial neoplasia. Additionally, increased CpG site methylation was found in low levels in most of the five host nuclear genes studied, including 5/5 TERT, 1/4 DAPK1, 2/5 RARB, MAL, and CADM1.[69] Furthermore, 1/3 of CpG sites in mitochondrial DNA were associated with increased methylation in CIN3+.[69] Thus, a correlation exists between CIN3+ and increased methylation of CpG sites in the HPV16 L1 open reading frame.[69] This could be a potential biomarker for future screens of cancerous and precancerous cervical disease.[69]
Current therapeutics
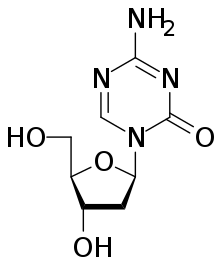
Drugs that specifically target the inverted methylation pattern of cancerous cells include the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine. This hypomethylating agent has been used to treat myelodysplastic syndrome, a blood cancer produced by abnormal bone marrow stem cells.[3] An inhibitor for all three types of active DNA methyltransferases, 5-azaC, previously thought to be highly toxic for human trials, proves to be an effective therapeutics in clinical trials when apply in low dosage, reducing progression of myelodysplastic syndrome to leukaemia, and increasing survival rate of patients with cancer.[70]
Histone deacetylase (HDAC) inhibitors have undergone many clinical trials. With significant efficacy in treating T cell lymphoma, two HDAC inhibitors, vorinostat and romidepsin, have recently been approved by the Food and Drug Administration.[71][72] However, since these HDAC inhibitors alter the acetylation state of many proteins in addition to the histone of interest, knowledge of the underlying mechanism at the molecular level of patient response is required to enhance the efficiency of using such inhibitors as treatment.[9] A histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA) has had some success in clinical trials.[73] Epigenetic therapeutics are taken in combination and concurrently with traditional cancer treatments, such as chemotherapy and immunotherapy. There is much crosstalk between CpG island methylation and histone modifications. Treatment with HDAC inhibitors has been found to promote gene reactivation after DNA methyl-transferases inhibitors have repressed transcription.[74]
Tools for identification
Previously, epigenetic profiles were limited to individual genes under scrutiny by a particular research team. Recently, however, scientists have been moving toward a more genomic approach to determine an entire genomic profile for cancerous versus healthy cells.[2]
Popular approaches for measuring CpG methylation in cells include:
- Bisulfite sequencing
- Combined bisulfite restriction analysis (COBRA)
- MethyLight
- Pyrosequencing
- Restriction landmark genomic scanning
- Arbitrary primed PCR
- HELP assay (HpaII tiny fragment enrichment by ligation-mediated PCR)
- Chromatin immunoprecipitation ChIP-Chip using antibodies specific for methyl-CpG binding domain proteins
- Methylated DNA immunoprecipitation Methyl-DIP
- Gene-expression profiles via DNA microarray : comparing mRNA levels from cancer cell lines before and after treatment with a demethylating agent
Since bisulfite sequencing is considered the gold standard for measuring CpG methylation, when one of the other methods is used, results are usually confirmed using bisulfite sequencing[1]. Popular approaches for determining histone modification profiles in cancerous versus healthy cells include:[2]
- Mass spectrometry
- Chromatin Immunoprecipitation Assay
Diagnostic and prognostic uses
Researchers are hoping to identify specific epigenetic profiles of various types and subtypes of cancer with the goal of using these profiles as tools to diagnose individuals more specifically and accurately.[2] Since epigenetic profiles change, scientists would like to use the different epigenomic profiles to determine the stage of development or level of aggressiveness of a particular cancer in patients. For example, hypermethylation of the genes coding for Death-Associated Protein Kinase (DAPK), p16, and Epithelial Membrane Protein 3 (EMP3) have been linked to more aggressive forms of lung, colorectal, and brain cancers.[8] This type of knowledge can affect the way that doctors will diagnose and choose to treat their patients.
Another factor that will influence the treatment of patients is knowing how well they will respond to certain treatments. Personalized epigenomic profiles of cancerous cells can provide insight into this field. For example, MGMT is an enzyme that reverses the addition of alkyl groups to the nucleotide guanine.[75] Alkylating guanine, however, is the mechanism by which several chemotherapeutic drugs act in order to disrupt DNA and cause cell death.[76][77][78][79] Therefore, if the gene encoding MGMT in cancer cells is hypermethylated and in effect silenced or repressed, the chemotherapeutic drugs that act by methylating guanine will be more effective than in cancer cells that have a functional MGMT enzyme.
Epigenetic biomarkers can also be utilized as tools for molecular prognosis. In primary tumor and mediastinal lymph node biopsy samples, hypermethylation of both CDKN2A and CDH13 serves as the marker for increased risk of faster cancer relapse and higher death rate of patients.[80]
References
- ^ Novak, Kris (20 December 2004). "Epigenetics Changes in Cancer Cells". Medscape General Medicine. 6 (4): 17. PMC 1480584. PMID 15775844.
{{cite journal}}:|access-date=requires|url=(help) - ^ a b c d e f g h Esteller Manel (2007). "Cancer epigenomics: DNA methylomes and histone-modification maps". Nat Rev Genet. 8 (4): 286–98. doi:10.1038/nrg2005. PMID 17339880.
- ^ a b c Jones PA, Baylin SB (2002). "The fundamental role of epigenetic events in cancer". Nat Rev Genet. 3 (6): 415–28. doi:10.1038/nrg816. PMID 12042769.
- ^ De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. (2012). "DNA methylation screening identifies driver epigenetic events of cancer cell survival". Cancer Cell. 21 (5): 655–67. doi:10.1016/j.ccr.2012.03.045. PMC 3395886. PMID 22624715.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Herman JG, Baylin SB (2003). "Gene silencing in cancer in association with promoter hypermethylation". N Engl J Med. 349 (21): 2042–54. doi:10.1056/NEJMra023075. PMID 14627790.
- ^ a b c Feinberg AP, Tycko B (2004). "The history of cancer epigenetics". Nat Rev Cancer. 4 (2): 143–53. doi:10.1038/nrc1279. PMID 14732866.
- ^ a b c Egger G, Liang G, Aparicio A, Jones PA (2004). "Epigenetics in human disease and prospects for epigenetic therapy". Nature. 429 (6990): 457–63. doi:10.1038/nature02625. PMID 15164071.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Esteller M (2005). "Aberrant DNA methylation as a cancer-inducing mechanism". Annu Rev Pharmacol Toxicol. 45: 629–56. doi:10.1146/annurev.pharmtox.45.120403.095832. PMID 15822191.
- ^ a b c Baylin SB, Jones PA (2011). "A decade of exploring the cancer epigenome - biological and translational implications". Nat Rev Cancer. 11 (10): 726–34. doi:10.1038/nrc3130. PMC 3307543. PMID 21941284.
- ^ a b Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G; et al. (2005). "Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer". Nat Genet. 37 (4): 391–400. doi:10.1038/ng1531. PMID 15765097.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A; et al. (2009). "Histone H4 lysine 16 acetylation regulates cellular lifespan". Nature. 459 (7248): 802–7. doi:10.1038/nature08085. PMC 2702157. PMID 19516333.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C; et al. (2006). "The Polycomb group protein EZH2 directly controls DNA methylation". Nature. 439 (7078): 871–4. doi:10.1038/nature04431. PMID 16357870.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Richon VM, Sandhoff TW, Rifkind RA, Marks PA (2000). "Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation". Proc Natl Acad Sci U S A. 97 (18): 10014–9. doi:10.1073/pnas.180316197. PMC 27656. PMID 10954755.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ jobs (2012-08-08). "Cancer research: Open ambition : Nature News & Comment". Nature. Retrieved 2013-01-19.
- ^ a b Soto-Reyes E, Recillas-Targa F (2010). "Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines". Oncogene. 29 (15): 2217–27. doi:10.1038/onc.2009.509. PMID 20101205.
- ^ Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M; et al. (2006). "A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition". Nat Genet. 38 (5): 566–9. doi:10.1038/ng1773. PMID 16642021.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Van Attikum H, Gasser SM (2009). "Crosstalk between histone modifications during the DNA damage response". Trends Cell Biology. 19 (5): 207–17. doi:10.1016/j.tcb.2009.03.001. PMID 19342239.
- ^ Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA; et al. (2006). "Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells". Cancer Cell. 9 (6): 435–43. doi:10.1016/j.ccr.2006.04.020. PMID 16766263.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F; et al. (2007). "Genetic unmasking of an epigenetically silenced microRNA in human cancer cells". Cancer Res. 67 (4): 1424–9. doi:10.1158/0008-5472.CAN-06-4218. PMID 17308079.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Soto-Reyes E, González-Barrios R, Cisneros-Soberanis F, Herrera-Goepfert R, Pérez V, Cantú D; et al. (2012). "Disruption of CTCF at the miR-125b1 locus in gynecological cancers". BMC Cancer. 12: 40. doi:10.1186/1471-2407-12-40. PMC 3297514. PMID 22277129.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c Bernstein C, Prasad AR, Nfonsam V, Bernstein H. (2013). DNA Damage, DNA Repair and Cancer, New Research Directions in DNA Repair, Prof. Clark Chen (Ed.), ISBN: 978-953-51-1114-6, InTech, http://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-damage-dna-repair-and-cancer
- ^ a b O'Hagan HM, Mohammad HP, Baylin SB. (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island" PLoS Genet 4(8) e1000155. doi:10.1371/journal.pgen.1000155 PMID 18704159
- ^ a b Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV (2007). DNA damage, homology-directed repair, and DNA methylation" PLoS Genet 3(7) e110. doi:10.1371/journal.pgen.0030110 PMID 17616978
- ^ Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010). Hereditary and familial colon cancer" Gastroenterology 138(6) 2044-2058. doi: 10.1053/j.gastro.2010.01.054. PMID 20420945
- ^ Truninger K, Menigatti M, Luz J; et al. (May 2005). "Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer". Gastroenterology. 128 (5): 1160–71. doi:10.1053/j.gastro.2005.01.056. PMID 15887099.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Valeri N, Gasparini P, Fabbri M; et al. (April 2010). "Modulation of mismatch repair and genomic stability by miR-155". Proc. Natl. Acad. Sci. U.S.A. 107 (15): 6982–7. Bibcode:2010PNAS..107.6982V. doi:10.1073/pnas.1002472107. PMC 2872463. PMID 20351277.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b c Zhang W, Zhang J, Hoadley K; et al. (June 2012). "miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression". Neuro-oncology. 14 (6): 712–9. doi:10.1093/neuonc/nos089. PMC 3367855. PMID 22570426.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Spiegl-Kreinecker S, Pirker C, Filipits M; et al. (January 2010). "O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients". Neuro-oncology. 12 (1): 28–36. doi:10.1093/neuonc/nop003. PMC 2940563. PMID 20150365.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Sgarra R, Rustighi A, Tessari MA; et al. (September 2004). "Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer". FEBS Lett. 574 (1–3): 1–8. doi:10.1016/j.febslet.2004.08.013. PMID 15358530.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Xu Y, Sumter TF, Bhattacharya R; et al. (May 2004). "The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia". Cancer Res. 64 (10): 3371–5. doi:10.1158/0008-5472.CAN-04-0044. PMID 15150086.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Baldassarre G, Battista S, Belletti B; et al. (April 2003). "Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma". Mol. Cell. Biol. 23 (7): 2225–38. doi:10.1128/MCB.23.7.2225-2238.2003. PMC 150734. PMID 12640109.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Borrmann L, Schwanbeck R, Heyduk T; et al. (December 2003). "High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity". Nucleic Acids Res. 31 (23): 6841–51. doi:10.1093/nar/gkg884. PMC 290254. PMID 14627817.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b c d e Facista A, Nguyen H, Lewis C, Prasad AR, Ramsey L, Zaitlin B, Nfonsam V, Krouse RS, Bernstein H, Payne CM, Stern S, Oatman N, Banerjee B, Bernstein C. (2012). Deficient expression of DNA repair enzymes in early progression to sporadic colon cancer. Genome Integr 3(1) 3. doi: 10.1186/2041-9414-3-3. PMID 22494821 http://www.ncbi.nlm.nih.gov/pubmed/22494821
- ^ Palmieri D, D'Angelo D, Valentino T; et al. (August 2012). "Downregulation of HMGA-targeting microRNAs has a critical role in human pituitary tumorigenesis". Oncogene. 31 (34): 3857–65. doi:10.1038/onc.2011.557. PMID 22139073.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Malumbres M (2013). "miRNAs and cancer: an epigenetics view". Mol. Aspects Med. 34 (4): 863–74. doi:10.1016/j.mam.2012.06.005. PMID 22771542.
- ^ a b Schnekenburger M, Diederich M (March 2012). "Epigenetics Offer New Horizons for Colorectal Cancer Prevention". Curr Colorectal Cancer Rep. 8 (1): 66–81. doi:10.1007/s11888-011-0116-z. PMC 3277709. PMID 22389639.
- ^ Sampath D, Liu C, Vasan K; et al. (February 2012). "Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia". Blood. 119 (5): 1162–72. doi:10.1182/blood-2011-05-351510. PMC 3277352. PMID 22096249.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Son D.j., Kumar S., Takabe W., Kim C.W., Ni C.W., Alberts-Grill N., Jang I.H., Kim S., Kim W., Kang S.W., Baker A.H., Seo J.W., Ferrara K.W., and Jo H. (2013). The atypical mechnosenstitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nature Communications. 4:3000.
- ^ Wan G, Mathur R, Hu X, Zhang X, Lu X (September 2011). "miRNA response to DNA damage". Trends Biochem. Sci. 36 (9): 478–84. doi:10.1016/j.tibs.2011.06.002. PMC 3532742. PMID 21741842.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E (2014). "MicroRNAs in the DNA Damage/Repair Network and Cancer". Int J Genomics. 2014: 820248. doi:10.1155/2014/820248. PMC 3926391. PMID 24616890.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Tuna M, Amos CI (November 2013). "Genomic sequencing in cancer". Cancer Lett. 340 (2): 161–70. doi:10.1016/j.canlet.2012.11.004. PMID 23178448.
- ^ Roach JC, Glusman G, Smit AF; et al. (April 2010). "Analysis of genetic inheritance in a family quartet by whole-genome sequencing". Science. 328 (5978): 636–9. doi:10.1126/science.1186802. PMC 3037280. PMID 20220176.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Campbell CD, Chong JX, Malig M; et al. (November 2012). "Estimating the human mutation rate using autozygosity in a founder population". Nat. Genet. 44 (11): 1277–81. doi:10.1038/ng.2418. PMC 3483378. PMID 23001126.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM (April 1997). "Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2". Proc. Natl. Acad. Sci. U.S.A. 94 (7): 3122–7. doi:10.1073/pnas.94.7.3122. PMC 20332. PMID 9096356.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM (December 2006). "Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6". Carcinogenesis. 27 (12): 2402–8. doi:10.1093/carcin/bgl079. PMC 2612936. PMID 16728433.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tutt AN, van Oostrom CT, Ross GM, van Steeg H, Ashworth A (March 2002). "Disruption of Brca2 increases the spontaneous mutation rate in vivo: synergism with ionizing radiation". EMBO Rep. 3 (3): 255–60. doi:10.1093/embo-reports/kvf037. PMC 1084010. PMID 11850397.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fearon, ER (1997). "Human cancer syndromes: Clues to the origin and nature of cancer". Science. 278 (5340): 1043–1050. doi:10.1126/science.278.5340.1043. PMID 9353177.
- ^ Halford, S; Rowan, A; Sawyer, E; Talbot, I; Tomlinson, I (2005). "O6-methylguanine methyltransferase in colorectal cancers: Detection of mutations, loss of expression, and weak association with G:C>A:T transitions". Gut. 54 (6): 797–802. doi:10.1136/gut.2004.059535. PMC 1774551. PMID 15888787.
- ^ Truninger, Kaspar; Menigatti, Mirco; Luz, Judith; Russell, Anna; Haider, Ritva; Gebbers, Jan-Olaf; Bannwart, Fridolin; Yurtsever, Hueseyin; Neuweiler, Joerg; Riehle, Hans-Martin; Cattaruzza, Maria Sofia; Heinimann, Karl; Schär, Primo; Jiricny, Josef; Marra, Giancarlo (2005). "Immunohistochemical Analysis Reveals High Frequency of PMS2 Defects in Colorectal Cancer". Gastroenterology. 128 (5): 1160–1171. doi:10.1053/j.gastro.2005.01.056. PMID 15887099.
- ^ Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; Costinean, S.; Sandhu, S. K.; Nuovo, G. J.; Alder, H.; Gafa, R.; Calore, F.; Ferracin, M.; Lanza, G.; Volinia, S.; Negrini, M.; McIlhatton, M. A.; Amadori, D.; Fishel, R.; Croce, C. M. (2010). "Modulation of mismatch repair and genomic stability by miR-155". Proceedings of the National Academy of Sciences. 107 (15): 6982–6987. doi:10.1073/pnas.1002472107. PMC 2872463. PMID 20351277.
- ^ Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JP (2005). "MGMT promoter methylation and field defect in sporadic colorectal cancer". J. Natl. Cancer Inst. 97 (18): 1330–8. doi:10.1093/jnci/dji275. PMID 16174854.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Cite error: The named reference
Lee KH 2011was invoked but never defined (see the help page). - ^ Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, Seruca R, Gaub MP, Legrain M, Collura A, Lascols O, Tiret E, Fléjou JF, Duval A (November 2010). "Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers". Gut. 59 (11): 1516–26. doi:10.1136/gut.2009.194787. PMID 20947886.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lee KH, Lee JS, Nam JH, Choi C, Lee MC, Park CS, Juhng SW, Lee JH (2011). "Promoter methylation status of hMLH1, hMSH2, and MGMT genes in colorectal cancer associated with adenoma-carcinoma sequence". Langenbecks Arch Surg. 396 (7): 1017–26. doi:10.1007/s00423-011-0812-9. PMID 21706233.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Paluszczak J, Misiak P, Wierzbicka M, Woźniak A, Baer-Dubowska W (February 2011). "Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa". Oral Oncol. 47 (2): 104–7. doi:10.1016/j.oraloncology.2010.11.006. PMID 21147548.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zuo C, Zhang H, Spencer HJ, Vural E, Suen JY, Schichman SA, Smoller BR, Kokoska MS, Fan CY (October 2009). "Increased microsatellite instability and epigenetic inactivation of the hMLH1 gene in head and neck squamous cell carcinoma". Otolaryngol Head Neck Surg. 141 (4): 484–90. doi:10.1016/j.otohns.2009.07.007. PMID 19786217.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tawfik HM, El-Maqsoud NM, Hak BH, El-Sherbiny YM (2011). "Head and neck squamous cell carcinoma: mismatch repair immunohistochemistry and promoter hypermethylation of hMLH1 gene". Am J Otolaryngol. 32 (6): 528–36. doi:10.1016/j.amjoto.2010.11.005. PMID 21353335.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ (November 2009). "Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions". Hum. Pathol. 40 (11): 1534–42. doi:10.1016/j.humpath.2009.01.029. PMID 19695681.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G, Wani R, Wani K (2012). "Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley". Asian Pac. J. Cancer Prev. 13 (8): 4177–81. doi:10.7314/APJCP.2012.13.8.4177. PMID 23098428.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Agarwal A, Polineni R, Hussein Z, Vigoda I, Bhagat TD, Bhattacharyya S, Maitra A, Verma A (2012). "Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma". Int J Clin Exp Pathol. 5 (5): 382–96. PMC 3396065. PMID 22808291.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html
- ^ Xinrong Chen and Tao Chen (2011). Roles of MicroRNA in DNA Damage and Repair, DNA Repair, Dr. Inna Kruman (Ed.), ISBN 978-953-307-697-3, InTech, Available from: http://www.intechopen.com/books/dnarepair/roles-of-microrna-in-dna-damage-and-repair
- ^ Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM. (2013). MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 19(2):230-42. doi: 10.1261/rna.034926.112. PMID 23249749 http://www.ncbi.nlm.nih.gov/pubmed/23249749
- ^ Chaisaingmongkol J, Popanda O, Warta R, Dyckhoff G, Herpel E, Geiselhart L, Claus R, Lasitschka F, Campos B, Oakes CC, Bermejo JL, Herold-Mende C, Plass C, Schmezer P. (2012). Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma. Oncogene 31(49):5108–16. doi: 10.1038/onc.2011.660. PMID 22286769
- ^ Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010). "ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks". Cell. 141 (6): 970–81. doi:10.1016/j.cell.2010.04.038. PMC 2920610. PMID 20550933.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Morano A, Angrisano T, Russo G, Landi R, Pezone A, Bartollino S, Zuchegna C, Babbio F, Bonapace IM, Allen B, Muller MT, Chiariotti L, Gottesman ME, Porcellini A, Avvedimento EV (2014). "Targeted DNA methylation by homology-directed repair in mammalian cells. Transcription reshapes methylation on the repaired gene". Nucleic Acids Res. 42 (2): 804–21. doi:10.1093/nar/gkt920. PMC 3902918. PMID 24137009.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Collins CC, Volik SV, Lapuk AV, Wang Y, Gout PW, Wu C; et al. (2012). "Next generation sequencing of prostate cancer from a patient identifies a deficiency of methylthioadenosine phosphorylase, an exploitable tumor target". Mol Cancer Ther. 11 (3): 775–83. doi:10.1158/1535-7163.MCT-11-0826. PMID 22252602.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b c Gurel B, Iwata T, Koh CM, Yegnasubramanian S, Nelson WG, De Marzo AM (2008). "Molecular alterations in prostate cancer as diagnostic, prognostic, and therapeutic targets". Adv Anat Pathol. 15 (6): 319–31. doi:10.1097/PAP.0b013e31818a5c19. PMC 3214657. PMID 18948763.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f g h Sun C, Reimers LL, Burk RD (2011). "Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer". Gynecol Oncol. 121 (1): 59–63. doi:10.1016/j.ygyno.2011.01.013. PMC 3062667. PMID 21306759.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A; et al. (2009). "Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study". Lancet Oncol. 10 (3): 223–32. doi:10.1016/S1470-2045(09)70003-8. PMID 19230772.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Duvic M, Talpur R, Ni X, Zhang C, Hazarika P, Kelly C; et al. (2007). "Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL)". Blood. 109 (1): 31–9. doi:10.1182/blood-2006-06-025999. PMC 1785068. PMID 16960145.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, Parker S; et al. (2007). "Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma". J Clin Oncol. 25 (21): 3109–15. doi:10.1200/JCO.2006.10.2434. PMID 17577020.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ C. David Allis (2007). Epigenetics. CSHL Press. ISBN 978-0-87969-724-2. Retrieved 17 May 2012.
- ^ Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB (1999). "Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer". Nat Genet. 21 (1): 103–7. doi:10.1038/5047. PMID 9916800.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Esteller M, Herman JG (2004). "Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer". Oncogene. 23 (1): 1–8. doi:10.1038/sj.onc.1207316. PMID 14712205.
- ^ Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V; et al. (2000). "Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents". N Engl J Med. 343 (19): 1350–4. doi:10.1056/NEJM200011093431901. PMID 11070098.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M; et al. (2005). "MGMT gene silencing and benefit from temozolomide in glioblastoma". N Engl J Med. 352 (10): 997–1003. doi:10.1056/NEJMoa043331. PMID 15758010.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B; et al. (2002). "Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma". J Natl Cancer Inst. 94 (1): 26–32. PMID 11773279.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Glasspool RM, Teodoridis JM, Brown R (2006). "Epigenetics as a mechanism driving polygenic clinical drug resistance". Br J Cancer. 94 (8): 1087–92. doi:10.1038/sj.bjc.6603024. PMC 2361257. PMID 16495912.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S; et al. (2008). "DNA methylation markers and early recurrence in stage I lung cancer". N Engl J Med. 358 (11): 1118–28. doi:10.1056/NEJMoa0706550. PMID 18337602.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link)
