Chromosomal inversion


An inversion is a chromosome rearrangement in which a segment of a chromosome is reversed end-to-end. An inversion occurs when a single chromosome undergoes breakage and rearrangement within itself. Inversions are of two types: paracentric and pericentric.
Paracentric inversions do not include the centromere, and both breaks occur in one arm of the chromosome. Pericentric inversions include the centromere, and there is a break point in each arm.
Cytogenetic techniques may be able to detect inversions, or inversions may be inferred from genetic analysis. Nevertheless, in most species, small inversions go undetected. In insects with polytene chromosomes (for example, Drosophila), preparations of larval salivary gland chromosomes allow inversions to be seen when they are heterozygous. This useful characteristic of polytene chromosomes was first noted by Theophilus Shickel Painter in 1933.[1]
Inversions usually do not cause any abnormalities in carriers, as long as the rearrangement is balanced, with no extra or missing DNA. However, in individuals which are heterozygous for an inversion, there is an increased production of abnormal chromatids (this occurs when crossing-over occurs within the span of the inversion). This leads to lowered fertility, due to production of unbalanced gametes. An inversion does not involve a loss of genetic information, but simply rearranges the linear gene sequence.
Human families that may be carriers of inversions may be offered genetic counseling and genetic testing.[2]
In the species Drosophila subobscura, researchers have been able to track global climate change by measuring the magnitude and directional shift in chromosome inversion frequencies, relative to temperatures at specific global sites.[3]
An example of chromosomal Inversion in organisms is demonstrated in the insect, Coelopa frigida. This particular species of Coelopa have a variation of chromosomal inversions that allow the species to create a series of physical differences. Individual C. frigida that are larger do not undergo a chromosomal inversion, whereas individuals that are smaller do undergo a chromosomal inversion.
Nomenclature

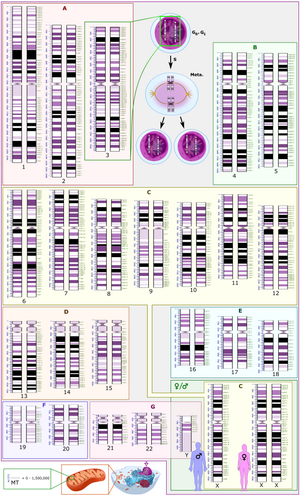
The International System for Human Cytogenomic Nomenclature (ISCN) is an international standard for human chromosome nomenclature, which includes band names, symbols and abbreviated terms used in the description of human chromosome and chromosome abnormalities. Abbreviations include inv for inversions.[5]
Notable cases
- Brenden Adams: former holder of the Guinness World Record for tallest teenager. His height is caused by an inversion of chromosome 12.
References
- ^ Painter TS (1933). "A new method for the study of chromosome rearrangements and the plotting of chromosome maps". Science. 78 (2034): 585–6. Bibcode:1933Sci....78..585P. doi:10.1126/science.78.2034.585. PMID 17801695.
- ^ Gardner, R.J.M; Sutherland, Grant R.; Shaffer, Lisa G. (2011). "9 Inversions". Chromosome Abnormalities and Genetic Counseling (4th ed.). Oxford University Press. pp. 161–182. ISBN 978-0-19-974915-7.
- ^ "Global Genetic Change Tracks Global Climate Warming in Drosophila subobscura". Science. 313 (5794):1773–5): 1773–1775. 2006. Bibcode:2006Sci...313.1773B. doi:10.1126/science.1131002. PMID 16946033. S2CID 7525234.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Warrender JD, Moorman AV, Lord P (2019). "A fully computational and reasonable representation for karyotypes". Bioinformatics. 35 (24): 5264–5270. doi:10.1093/bioinformatics/btz440. PMC 6954653. PMID 31228194.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- "This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/)" - ^ "ISCN Symbols and Abbreviated Terms". Coriell Institute for Medical Research. Retrieved 2022-10-27.
Further reading
- Lehtonen S, Myllys L, Huttunen S (2009). "Phylogenetic analysis of non-coding plastid DNAthtjtdjj in the presence of short inversions" (PDF-preview). Phytotaxa. 1: 3–20. doi:10.11646/phytotaxa.1.1.2.
