Cyanine: Difference between revisions
m robot Modifying: de:Cyanine |
Cy5 ozone susceptibility |
||
| Line 33: | Line 33: | ||
The [[scanners]] actually use different [[laser]] emission wavelengths (typically 532 [[Metre#SI prefixed forms of metre|nm]] and 635 [[Metre#SI prefixed forms of metre|nm]]) and filter wavelengths (550-600 [[Metre#SI prefixed forms of metre|nm]] and 655-695 [[Metre#SI prefixed forms of metre|nm]]) to avoid background contamination. They are thus able to easily distinguish between two samples when one sample has been labeled with Cy3 and the other labeled with Cy5. They are also able to quantitate the amount of labeling in either sample. |
The [[scanners]] actually use different [[laser]] emission wavelengths (typically 532 [[Metre#SI prefixed forms of metre|nm]] and 635 [[Metre#SI prefixed forms of metre|nm]]) and filter wavelengths (550-600 [[Metre#SI prefixed forms of metre|nm]] and 655-695 [[Metre#SI prefixed forms of metre|nm]]) to avoid background contamination. They are thus able to easily distinguish between two samples when one sample has been labeled with Cy3 and the other labeled with Cy5. They are also able to quantitate the amount of labeling in either sample. |
||
=== Cy5 ozone susceptibility === |
|||
In 2003 researchers at Inpharmatics and Agilent reported in the journal [[Analytical Chemistry (journal)]] that Cy5 is suspectibile to environmental ozone. Exposures to ozone levels above 5-10 ppb for 10-30 seconds were reported to decrease the reproducibility of Cy5 microarrays. Much higher levels of ozone (>100 ppb) were required to observe an affect in Cy3 <ref name=fare>Fare TL, Coffey EM, Hongyue D, et.al. Effects of Atmospheric Ozone on Microarray Data Quality. Analytical Chemistry. 2003;75:4672-4675. [http://pubs.acs.org/ncw/2003/articles/ac034241b.pdf]</ref>. |
|||
=== Applications === |
=== Applications === |
||
Revision as of 18:55, 3 March 2007

Cyanine is a non-systematic name of a synthetic dye family with the common molecular formula:
ArN+=CH[CH=CH]n=NAr
where two quaternized nitrogens are joined by a poly-methine chain[1]. Both nitrogens are each independently part of a heteroaromatic moiety, such as imidazole, pyridine, pyrrole, quinoline, thiazole, etc.
Cyanines were first synthesized over a century ago, and there are a large number reported in the literature.
Uses
They have many uses as fluorescent dyes. Depending on the structure they cover the spectrum from IR to UV.
They were originally used, and still are, to increase the sensitivity range of photographic emulsions, i.e. to increase the range of wavelengths which will form an image on the film.
They are used in CD-R and DVD-R media. The ones used are mostly green or light blue in color, and are chemically unstable. This makes cyanine discs unsuitable for archival CD and DVD use, as they can fade and become unreadable in a few years.
Cy3 and Cy5
Cy3 and Cy5 are dyes used in comparative genomic hybridization, and in gene chips which are used in transcriptomics. They are also used to label proteins for various studies including proteomics. Cy3 is a green colour and Cy5 is red.
Cy3 and Cy5 are reactive watersoluble fluorescent dyes of the cyanine dye family. They are usually synthesized with reactive groups on either one or both of the nitrogen side chains, so that they can be chemically cross-linked to either nucleic acids or protein molecules. Labeling is done for visualization and quantification purposes.
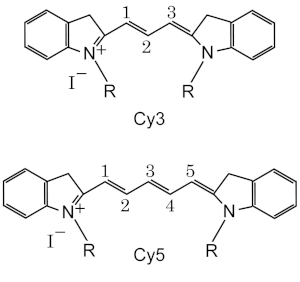
Nomenclature and Structure
Standard chemical names specify exactly the chemical structure of the molecule. The Cy3 and Cy5 nomenclature was first proposed by Ernst, et al.[1] in 1989, and is non-standard, since it gives no hint of their chemical strucutres. In the original paper the number designated the count of the methines (as shown), and the side chains were unspecified. Thus various structures are designated by Cy3 and Cy5 in the literature.
The R groups do not have to be identical. In the dyes as used they are short aliphatic chains one or both of which ends in a highly reactive moieties such as N-hydroxysuccinimide or maleimide.
Spectral characteristics
Cy3 is excited maximally at 550 nm and emits maximally at 570 nm, in the green part of the spectrum; quantum yield is 0.15; FW=766.
Cy5 is excited maximally at 649 nm and emits maximally at 670 nm, in the red part of the spectrum; quantum yield is 0.28. FW=792.
The scanners actually use different laser emission wavelengths (typically 532 nm and 635 nm) and filter wavelengths (550-600 nm and 655-695 nm) to avoid background contamination. They are thus able to easily distinguish between two samples when one sample has been labeled with Cy3 and the other labeled with Cy5. They are also able to quantitate the amount of labeling in either sample.
Cy5 ozone susceptibility
In 2003 researchers at Inpharmatics and Agilent reported in the journal Analytical Chemistry (journal) that Cy5 is suspectibile to environmental ozone. Exposures to ozone levels above 5-10 ppb for 10-30 seconds were reported to decrease the reproducibility of Cy5 microarrays. Much higher levels of ozone (>100 ppb) were required to observe an affect in Cy3 [2].
Applications
Nucleic acid labeling
In microarray experiments DNA or RNA is labeled with either Cy3 or Cy5 that has been synthesized to carry the reactive group N-hydroxysuccinimide (NHS). Since, N-hydroxysuccinimide reacts readily only with aliphatic amine groups, which nucleic acids lack, nucleotides have to be modified with aminoallyl groups. This is done through incorporating aminoallyl-modified nucleotides during polymerase reactions. A good ratio is a label every 60 bases such that the labels are not too close to each other, thus resulting in quenching effects.
Protein labeling
For protein labeling, Cy3 and Cy5 dyes usually bear maleimide reactive groups instead*. The maleimide group allows conjugation of the fluorescent dye to the sulfhydryl group of cysteine residues. Cysteins can be added and removed from the protein domain of interest via PCR mutagenesis.
Cy5, is sensitive to the electronic environment it resides in. Changes in the conformation of the protein it is attached to will produce an enhancement or quenching of the emission. The rate of this change can be measured to determine enzyme kinetic parameters. The dyes can be used for similar purposes in FRET experiments.
Cy3 and Cy5 are used in proteomics experiments so that samples from two sources can be mixed and run together thorough the separation process[3]. This eliminates variations due to differing experimental conditions that are inevitable if the samples were run separately. These variations make it extremely difficult, if not impossible, to use computers to automate the acquisition of the data after the separation is complete. Using these dyes make makes the automation trivial.
References
- ^ a b Ernst LA, Gupta RK, Mujumdar RB, Waggoner AS. Cyanine dye labeling reagents for sulfhydryl groups. Cytometry. 1989;10(1):3-10. PMID: 2917472
- ^ Fare TL, Coffey EM, Hongyue D, et.al. Effects of Atmospheric Ozone on Microarray Data Quality. Analytical Chemistry. 2003;75:4672-4675. [1]
- ^ Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071-7. PMID: 9420172
