Endoplasmic reticulum: Difference between revisions
| Line 5: | Line 5: | ||
The '''endoplasmic reticulum''' ('''ER''') is an [[organelle]] of [[cell (biology)|cells]] in [[eukaryote|eukaryotic organisms]] that forms an interconnected network of membrane vesicles. According to the structure, the endoplasmic reticulum is classified into two types, '''rough endoplasmic reticulum''' '''(RER)''' and '''smooth endoplasmic reticulum''' '''(SER)'''. The rough endoplasmic reticulum is studded with ''ribosomes'' on the ''cytosolic'' face. These are the sites of ''protein synthesis''. The rough endoplasmic reticulum is predominantly found in ''[[Hepatocyte|hepatocytes]]'' where protein synthesis occurs actively. The smooth endoplasmic reticulum is a smooth network without the ribosomes. The smooth endoplasmic reticulum is concerned with ''lipid metabolism'', ''carbohydrate metabolism'', and ''detoxification''. The smooth endoplasmic reticulum is abundant in mammalian liver and gonad cells. The lacey membranes of the endoplasmic reticulum were first seen in 1945 by [[Keith R. Porter]], [[Albert Claude]], and Ernest F. Fullam.<ref name="FirstER">{{cite journal| author=Porter KR, Claude A, Fullam EF| year=1945| month=March| title=A study of tissue culture cells by electron microscopy| journal=J Exp Med.| volume=81| pages=233–246| doi=10.1084/jem.81.3.233| pmid=19871454| issue=3| pmc=2135493}}</ref> |
The '''endoplasmic reticulum''' ('''ER''') is an [[organelle]] of [[cell (biology)|cells]] in [[eukaryote|eukaryotic organisms]] that forms an interconnected network of membrane vesicles. According to the structure, the endoplasmic reticulum is classified into two types, '''rough endoplasmic reticulum''' '''(RER)''' and '''smooth endoplasmic reticulum''' '''(SER)'''. The rough endoplasmic reticulum is studded with ''ribosomes'' on the ''cytosolic'' face. These are the sites of ''protein synthesis''. The rough endoplasmic reticulum is predominantly found in ''[[Hepatocyte|hepatocytes]]'' where protein synthesis occurs actively. The smooth endoplasmic reticulum is a smooth network without the ribosomes. The smooth endoplasmic reticulum is concerned with ''lipid metabolism'', ''carbohydrate metabolism'', and ''detoxification''. The smooth endoplasmic reticulum is abundant in mammalian liver and gonad cells. The lacey membranes of the endoplasmic reticulum were first seen in 1945 by [[Keith R. Porter]], [[Albert Claude]], and Ernest F. Fullam.<ref name="FirstER">{{cite journal| author=Porter KR, Claude A, Fullam EF| year=1945| month=March| title=A study of tissue culture cells by electron microscopy| journal=J Exp Med.| volume=81| pages=233–246| doi=10.1084/jem.81.3.233| pmid=19871454| issue=3| pmc=2135493}}</ref> |
||
== |
==Strucdick== |
||
[[Image:nucleus ER golgi.svg|thumb|300px|'''1''' [[Cell nucleus|Nucleus]]{{nbsp|2}} |
[[Image:nucleus ER golgi.svg|thumb|300px|'''1''' [[Cell nucleus|Nucleus]]{{nbsp|2}} |
||
'''2''' [[Nuclear pore]]{{nbsp|2}} |
'''2''' [[Nuclear pore]]{{nbsp|2}} |
||
Revision as of 15:20, 9 October 2013
This article may be too technical for most readers to understand. (September 2013) |

The endoplasmic reticulum (ER) is an organelle of cells in eukaryotic organisms that forms an interconnected network of membrane vesicles. According to the structure, the endoplasmic reticulum is classified into two types, rough endoplasmic reticulum (RER) and smooth endoplasmic reticulum (SER). The rough endoplasmic reticulum is studded with ribosomes on the cytosolic face. These are the sites of protein synthesis. The rough endoplasmic reticulum is predominantly found in hepatocytes where protein synthesis occurs actively. The smooth endoplasmic reticulum is a smooth network without the ribosomes. The smooth endoplasmic reticulum is concerned with lipid metabolism, carbohydrate metabolism, and detoxification. The smooth endoplasmic reticulum is abundant in mammalian liver and gonad cells. The lacey membranes of the endoplasmic reticulum were first seen in 1945 by Keith R. Porter, Albert Claude, and Ernest F. Fullam.[1]
Strucdick
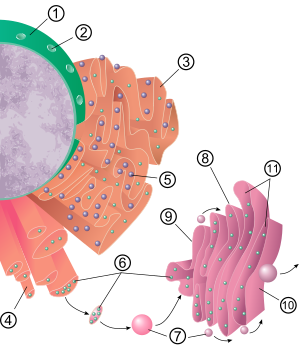

The general structure of the endoplasmic rectum is a membranous network of cisterns [disambiguation needed](sac-like structures) held together by the cytoskeleton. The phospholipid membrane encloses a space, the cisternal space (or lumen), which is continuous with the perinuclear space but separate from the cytosol. The functions of the endoplasmic reticulum vary greatly depending on cell type, cell function, and cell needs. The ER can even change over time in response to cell needs. The three most common varieties are called rough endoplasmic reticulum, smooth endoplasmic reticulum, and sarcoplasmic reticulum.
The quantity of RER and SER in a cell can slowly interchange from one type to the other, depending on changing metabolic needs. Transformation can include embedment of new proteins in membrane as well as structural changes. Massive changes may also occur in protein content without noticeable structural changes.
Rough endoplasmic reticulum

The surface of the rough endoplasmic reticulum (often abbreviated RER) is studded with protein-manufacturing ribosomes giving it a "rough" appearance (hence its name). The binding site of the ribosome on the rough endoplasmic reticulum is the translocon.[2] However, the ribosomes bound to it at any one time are not a stable part of this organelle's structure as they are constantly being bound and released from the membrane. A ribosome only binds to the RER once a specific protein-nucleic acid complex forms in the cytosol. This special complex forms when a free ribosome begins translating the mRNA of a protein destined for the secretory pathway.[3] The first 5-30 amino acids polymerized encode a signal peptide, a molecular message that is recognized and bound by a signal recognition particle (SRP). Translation pauses and the ribosome complex binds to the RER translocon where translation continues with the nascent protein forming into the RER lumen and/or membrane. The protein is processed in the ER lumen by an enzyme (a signal peptidase), which removes the signal peptide. Ribosomes at this point may be released back into the cytosol; however, non-translating ribosomes are also known to stay associated with translocons.[4]
The membrane of the rough endoplasmic reticulum forms large double membrane sheets that are located near, and continuous with, the outer layer of the nuclear envelope.[5] Although there is no continuous membrane between the endoplasmic reticulum and the Golgi apparatus, membrane-bound vesicles shuttle proteins between these two compartments.[6] Vesicles are surrounded by coating proteins called COPI and COPII. COPII targets vesicles to the golgi apparatus and COPI marks them to be brought back to the rough endoplasmic reticulum. The rough endoplasmic reticulum works in concert with the golgi complex to target new proteins to their proper destinations. A second method of transport out of the endoplasmic reticulum involves areas called membrane contact sites, where the membranes of the endoplasmic reticulum and other organelles are held closely together, allowing the transfer of lipids and other small molecules.[7][8]
The rough endoplasmic reticulum is key in multiple functions:
- Manufacture of lysosomal enzymes with a mannose-6-phosphate marker added in the cis-Golgi network[citation needed]
- Manufacture of secreted proteins, either secreted constitutively with no tag or secreted in a regulatory manner involving clathrin and paired basic amino acids in the signal peptide.
- Integral membrane proteins that stay embedded in the membrane as vesicles exit and bind to new membranes. Rab proteins are key in targeting the membrane; SNAP and SNARE proteins are key in the fusion event.
- Initial glycosylation as assembly continues. This is N-linked (O-linking occurs in the golgi).
Endoplasmic reticulum stress
Disturbances in redox regulation, calcium regulation, glucose deprivation, and viral infection [9] or the over-expression of proteins [10] can lead to endoplasmic reticulum stress (ER stress). It is a state in which the folding of proteins slows, leading to an increase in unfolded proteins. This stress is emerging as a potential cause of damage in hypoxia/ischemia, insulin resistance, and other disorders.
Smooth endoplasmic reticulum
The smooth endoplasmic reticulum (abbreviated SER) has functions in several metabolic processes. It synthesizes lipids, phospholipids, and steroids. Cells which secrete these products, such as those in the testes, ovaries, and skin oil glands have a great deal of smooth endoplasmic reticulum.[11] It also carries out the metabolism of carbohydrates, drug detoxification, attachment of receptors on cell membrane proteins, and steroid metabolism.[12] In muscle cells, it regulates calcium ion concentration. It is connected to the nuclear envelope. Smooth endoplasmic reticulum is found in a variety of cell types (both animal and plant), and it serves different functions in each. The smooth endoplasmic reticulum also contains the enzyme glucose-6-phosphatase, which converts glucose-6-phosphate to glucose, a step in gluconeogenesis. It consists of tubules that are located near the cell periphery. These tubes sometimes branch forming a network that is reticular in appearance.[5] In some cells, there are dilated areas like the sacs of rough endoplasmic reticulum. The network of smooth endoplasmic reticulum allows for an increased surface area to be devoted to the action or storage of key enzymes and the products of these enzymes.[citation needed]
Sarcoplasmic reticulum
The sarcoplasmic reticulum (SR), from the Greek sarx, ("flesh"), is smooth ER found in smooth and striated muscle. The only structural difference between this organelle and the smooth endoplasmic reticulum is the medley of proteins they have, both bound to their membranes and drifting within the confines of their lumens. This fundamental difference is indicative of their functions: The endoplasmic reticulum synthesizes molecules, while the sarcoplasmic reticulum stores and pumps calcium ions. The sarcoplasmic reticulum contains large stores of calcium, which it sequesters and then releases when the muscle cell is stimulated.[13][14] It plays a major role in excitation-contraction coupling.[citation needed]
Functions
The endoplasmic reticulum serves many general functions, including the facilitation of protein folding and the transport of synthesized proteins in sacs called cisterns.
Correct folding of newly made proteins is made possible by several endoplasmic reticulum chaperone proteins, including protein disulfide isomerase (PDI), ERp29, the Hsp70 family member BiP/Grp78, calnexin, calreticulin, and the peptidylpropyl isomerase family. Only properly folded proteins are transported from the rough ER to the Golgi apparatus.
Transport of proteins
Secretory proteins, mostly glycoproteins, are moved across the endoplasmic reticulum membrane. Proteins that are transported by the endoplasmic reticulum throughout the cell are marked with an address tag called a signal sequence. The N-terminus (one end) of a polypeptide chain (i.e., a protein) contains a few amino acids that work as an address tag, which are removed when the polypeptide reaches its destination. Proteins that are destined for places outside the endoplasmic reticulum are packed into transport vesicles and moved along the cytoskeleton toward their destination.
The endoplasmic reticulum is also part of a protein sorting pathway. It is, in essence, the transportation system of the eukaryotic cell. The majority of its resident proteins are retained within it through a retention motif. This motif is composed of four amino acids at the end of the protein sequence. The most common retention sequence is KDEL (lys-asp-glu-leu). However, variation on KDEL does occur and other sequences can also give rise to endoplasmic reticulum retention. It is not known whether such variation can lead to sub-ER localizations. There are three KDEL receptors in mammalian cells, and they have a very high degree of sequence identity. The functional differences between these receptors remain to be established. [citation needed]
See also
References
- ^ Porter KR, Claude A, Fullam EF (1945). "A study of tissue culture cells by electron microscopy". J Exp Med. 81 (3): 233–246. doi:10.1084/jem.81.3.233. PMC 2135493. PMID 19871454.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. (1992). "A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation". Cell. 71 (3): 489–503. PMID 1423609.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lodish, Harvey; et al. (2003). Molecular Cell Biology (5th ed.). W. H. Freeman. pp. 659–666. ISBN 0-7167-4366-3.
{{cite book}}: Explicit use of et al. in:|last2=(help) - ^ Seiser, R. M. (2000). "The Fate of Membrane-bound Ribosomes Following the Termination of Protein Synthesis". Journal of Biological Chemistry. 275 (43): 33820–33827. doi:10.1074/jbc.M004462200. ISSN 0021-9258.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Shibata, Yoko; Voeltz, Gia K.; Rapoport, Tom A. (2006). "Rough Sheets and Smooth Tubules". Cell. 126 (3): 435–439. doi:10.1016/j.cell.2006.07.019. ISSN 0092-8674. Cite error: The named reference "ShibataVoeltz2006" was defined multiple times with different content (see the help page).
- ^ Endoplasmic reticulum. (n.d.). McGraw-Hill Encyclopedia of Science and Technology. Retrieved September 13, 2006, from Answers.com Web site: http://www.answers.com/topic/endoplasmic-reticulum
- ^ Levine T (2004). "Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions". Trends Cell Biol. 14 (9): 483–90. doi:10.1016/j.tcb.2004.07.017. PMID 15350976.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Levine T, Loewen C (2006). "Inter-organelle membrane contact sites: through a glass, darkly". Curr. Opin. Cell Biol. 18 (4): 371–8. doi:10.1016/j.ceb.2006.06.011. PMID 16806880.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Xu, C (2005). "Endoplasmic Reticulum Stress: Cell Life and Death Decisions". J. Clin. Invest. 115 (10): 2656–2664. doi:10.1172/JCI26373. PMC 1236697. PMID 16200199.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kober L, Zehe C, Bode J (2012). "Development of a novel ER stress based selection system for the isolation of highly productive clones". Biotechnol. Bioeng. 109 (10): 2599–611. doi:10.1002/bit.24527. PMID 22510960.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "Functions of Smooth ER". University of Minnesota Duluth.
- ^ Maxfield FR, Wüstner D (2002). "Intracellular cholesterol transport". J. Clin. Invest. 110 (7): 891–8. doi:10.1172/JCI16500. PMC 151159. PMID 12370264.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Toyoshima C, Nakasako M, Nomura H, Ogawa H (2000). "Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution". Nature. 405 (6787): 647–55. doi:10.1038/35015017. PMID 10864315.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Medical Cell Biology 3rd/ed. Academic Press. p. 69.
