Homology (biology): Difference between revisions
Merged from Paralogy regions following December 2012 proposal; see Talk:Homology (biology)#Paralogy Regions |
No edit summary |
||
| Line 1: | Line 1: | ||
{{about||use of the term 'homologous' in reference to chromosomes|Homologous chromosomes|use of the term 'homologous' in reference to behaviors|Homology (psychology)}} |
{{about||use of the term 'homologous' in reference to chromosomes|Homologous chromosomes|use of the term 'homologous' in reference to behaviors|Homology (psychology)}}{{Other uses}} |
||
[[File:Homology vertebrates-en.svg|thumb|300px|The principle of homology: The biological derivation relationship (shown by colors) of the various bones in the forelimbs of four vertebrates is known as homology and was one of [[Charles Darwin]]’s arguments in favor of [[evolution]].]] |
[[File:Homology vertebrates-en.svg|thumb|300px|The principle of homology: The biological derivation relationship (shown by colors) of the various bones in the forelimbs of four vertebrates is known as homology and was one of [[Charles Darwin]]’s arguments in favor of [[evolution]].]] |
||
Revision as of 23:19, 26 July 2016
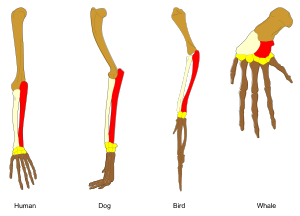
In the context of biology, homology is the existence of shared ancestry between a pair of structures, or genes, in different taxa.[1] A common example of homologous structures in evolutionary biology are the wings of bats and the arms of primates.[1] Evolutionary theory explains the existence of homologous structures adapted to different purposes as the result of descent with modification from a common ancestor.
In the context of sexual differentiation—the process of development of the differences between males and females from an undifferentiated fertilized egg — the male and female organs are homologous if they develop from the same embryonic tissue.[2] A typical example is the ovaries of female humans and the testicles of male humans.[2]
In the context of morphological differentiation, organs that developed in the same embryological manner from similar origins, such as from matching primordia in successive segments of the same organism, may be said to be homologous. Examples include the legs of a centipede, the maxillary palp and labial palp of an insect, and the spinous processes of successive vertebrae in a vertebral column. In contrast, a sesamoid bone such as the patella would not in any usual sense be regarded as homologous to a neighbouring skeletal bone such as the femur.
Etymology and Usage
The word homology, coined in about 1656, derives from the Greek ὁμόλογος homologos from ὁμός homos "same" and λόγος logos "relation". In biology, two things are homologous if they bear the same relationship to one another, such as a certain bone in various forms of the "hand". The alternative terms "homogeny" and "homogenous" were also used in the late 1800s and early 1900s. However, these terms are now archaic in biology, and the term "homogenous" is now generally found as a misspelling of the term "homogeneous" which refers to the uniformity of a mixture.[3][4][5][6]
Definition
Homology is the relationship between biological structures or sequences that are derived from a common ancestor. Homologous traits of organisms are therefore explained by descent from a common ancestor. The opposite of homologous organs are analogous organs which do similar jobs in two taxa that were not present in the last common ancestor but rather evolved separately. An example of an analogous trait would be the wings of bats and birds, which evolved independently in each lineage separately after diverging from ancestors with forelimbs not used as wings (terrestrial mammals and theropod dinosaurs, respectively).
It is important to distinguish between different hierarchical levels of homology in order to make informative biological comparisons. In the above example, the bird and bat wings are analogous as wings, but homologous as forelimbs because the organ served as a forearm (not a wing) in the last common ancestor of tetrapods.[7] Homology can also be described at the level of the gene. In genetics homology can refer to both the gene (DNA) and the corresponding protein product. It has been hypothesized that some behaviors might be homologous, based on either shared behavior across related taxa or common origins of the behavior in an individual’s development, though this remains controversial.
Anatomical homology

Evolutionary ancestry means that structures evolved from some structure in a common ancestor; for example, the wings of bats and the arms of primates are homologous in this sense. Developmental ancestry means that structures arose from the same tissue in embryonal development; the ovaries of female humans and the testicles of male humans are homologous in this sense.
Homology is different from analogy, which describes the relation between characters that are apparently similar yet phylogenetically independent. The wings of a maple seed and the wings of an albatross are analogous but not homologous (they both allow the organism to travel on the wind, but they didn't both develop from the same structure). Analogy is commonly also referred to as homoplasy, which is further distinguished into parallelism, reversal, and convergence.[8]
In evolutionary developmental biology (evo-devo), homology can also be partial. New structures can evolve through the combination of multiple developmental pathways, or parts of them. As a result, hybrid or mosaic structures can evolve that exhibit partial homologies. For example, certain compound leaves of flowering plants are partially homologous both to leaves and shoots because their development has evolved from a combination of leaf and shoot development.[9][10]
Determining homology
Systematists identify two forms of homology: primary homology is that initially conjectured by a researcher based on similar structure or anatomical connections, who states a hypothesis that two characters share an ancestry; secondary homology is implied by parsimony analysis, where a character that only occurs once on a tree is taken to be homologous.[11] As implied in this definition, many cladists consider homology to be synonymous with synapomorphy.
Homologous structures in other phyla
Introductory discussions of homology commonly limit themselves to the limbs of tetrapod vertebrates, occasionally touching on other structures, such as modified teeth as in whales and elephants. However, homologies provide the fundamental basis for all aspects of biological classification, although some of them may be highly counter-intuitive. For example, the embryonic body segments (somites) of different arthropod taxa (although the exact homology between the head appendages is still controversial).[12]
| Somite | Trilobite | Spider (Chelicerata) |
Centipede (Uniramia) |
Insect (Uniramia) |
Shrimp (Crustacea) |
|---|---|---|---|---|---|
| 1 | antennae | chelicerae (jaws and fangs) | antennae | antennae | 1st antennae |
| 2 | 1st legs | pedipalps | - | - | 2nd antennae |
| 3 | 2nd legs | 1st legs | mandibles | mandibles | mandibles (jaws) |
| 4 | 3rd legs | 2nd legs | 1st maxillae | 1st maxillae | 1st maxillae |
| 5 | 4th legs | 3rd legs | 2nd maxillae | 2nd maxillae | 2nd maxillae |
| 6 | 5th legs | 4th legs | collum (no legs) | 1st legs | 1st legs |
| 7 | 6th legs | - | 1st legs | 2nd legs | 2nd legs |
| 8 | 7th legs | - | 2nd legs | 3rd legs | 3rd legs |
| 9 | 8th legs | - | 3rd legs | - | 4th legs |
| 10 | 9th legs | - | 4th legs | - | 5th legs |
In many plants, defensive or storage structures are made by modifications of the development of primary leaves, stems, and roots.
| Primary organs | Defensive structures | Storage structures |
|---|---|---|
| Leaves | Spines | Swollen leaves (e.g. succulents) |
| Stems | Thorns | Tubers (e.g. potato), rhizomes (e.g. ginger), fleshy stems (e.g. cacti) |
| Roots | - | Root tubers (e.g. sweet potato), taproot (e.g. carrot) |
Sequence homology

Sequences are the amino acids for residues 120-180 of the proteins. Residues that are conserved across all sequences are highlighted in grey. Below the protein sequences is a key denoting conserved sequence (*), conservative mutations (:), semi-conservative mutations (.), and non-conservative mutations ( ).[13]
As with anatomical structures, homology between protein or DNA sequences is defined in terms of shared ancestry. Two segments of DNA can have shared ancestry because of either a speciation event (orthologs) or a duplication event (paralogs).[14]
Homology among proteins or DNA is typically inferred from their sequence similarity. Significant similarity is strong evidence that two sequences are related by divergent evolution of a common ancestor. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous.
The term "percent homology" is often incorrectly used to mean "sequence similarity". Sequence similarity (typically percent identity) is the observation, homology is the conclusion, since sequences are either homologous or not. As with anatomical structures, high sequence similarity might occur because of convergent evolution, or, as with shorter sequences, by chance. Such sequences are similar but not homologous.[citation needed] Sequence regions that are homologous are also called conserved. This is not to be confused with conservation in amino acid sequences in which the amino acid at a specific position has been substituted with a different one with functionally equivalent physicochemical properties.
Partial homology can occur where a segment of the sequences compared share descent, while the rest does not. For example, partial homology may result from a gene fusion event.
Orthology
Homologous sequences are orthologous if they are inferred to be descended from the same ancestral sequence separated by a speciation event: when a species diverges into two separate species, the copies of a single gene in the two resulting species are said to be orthologous. Orthologs, or orthologous genes, are genes in different species that originated by vertical descent from a single gene of the last common ancestor. The term "ortholog" was coined in 1970 by the molecular evolutionist Walter Fitch.[15]
For instance, the plant Flu regulatory protein is present both in Arabidopsis (multicellular higher plant) and Chlamydomonas (single cell green algae). The Chlamydomonas version is more complex: it crosses the membrane twice rather than once, contains additional domains and undergoes alternative splicing. However it can fully substitute the much simpler Arabidopsis protein, if transferred from algae to plant genome by means of gene engineering. Significant sequence similarity and shared functional domains indicate that these two genes are orthologous genes,[16] inherited from the shared ancestor.
Orthology is strictly defined in terms of ancestry. Given that the exact ancestry of genes in different organisms is difficult to ascertain due to gene duplication and genome rearrangement events, the strongest evidence that two similar genes are orthologous is usually found by carrying out phylogenetic analysis of the gene lineage. Orthologs often, but not always, have the same function.[17]
Orthologous sequences provide useful information in taxonomic classification and phylogenetic studies of organisms. The pattern of genetic divergence can be used to trace the relatedness of organisms. Two organisms that are very closely related are likely to display very similar DNA sequences between two orthologs. Conversely, an organism that is further removed evolutionarily from another organism is likely to display a greater divergence in the sequence of the orthologs being studied.
Databases of orthologous genes
Given their tremendous importance for biology and bioinformatics, orthologous genes have been organized in several specialized databases that provide tools to identify and analyze orthologous gene sequences. These resources employ approaches that can be generally classified into those that are based on all pairwise sequence comparisons (heuristic) and those that use phylogenetic methods. Sequence comparison methods were first pioneered by COGs,[18] now extended and automatically enhanced by the following databases:
- eggNOG[19]
- InParanoid[20] focuses on pairwise ortholog relationships
- OrthoDB[21] appreciates that the orthology concept is relative to different speciation points by providing a hierarchy of orthologs along the species tree.
- OrthoMCL[22]
- OMA
- Roundup[23]
- OrthoMaM[24] for mammals
- OrthologID[25]
- GreenPhylDB[26] for plants
Tree-based phylogenetic approaches aim to distinguish speciation from gene duplication events by comparing gene trees with species trees, as implemented in resources such as
A third category of hybrid approaches uses both heuristic and phylogenetic methods to construct clusters and determine trees, for example
Paralogy
Homologous sequences are paralogous if they were created by a duplication event within the genome. For gene duplication events, if a gene in an organism is duplicated to occupy two different positions in the same genome, then the two copies are paralogous.
Paralogous genes often belong to the same species, but this is not necessary: for example, the hemoglobin gene of humans and the myoglobin gene of chimpanzees are paralogs. Paralogs can be split into in-paralogs (paralogous pairs that arose after a speciation event) and out-paralogs (paralogous pairs that arose before a speciation event). Between-species out-paralogs are pairs of paralogs that exist between two organisms due to duplication before speciation, whereas within-species out-paralogs are pairs of paralogs that exist in the same organism, but whose duplication event happened before speciation. Paralogs typically have the same or similar function, but sometimes do not: due to lack of the original selective pressure upon one copy of the duplicated gene, this copy is free to mutate and acquire new functions.
Paralogous sequences provide useful and dramatic insight into some of the way genomes evolve. The genes encoding myoglobin and hemoglobin are considered to be ancient paralogs. Similarly, the four known classes of hemoglobins (hemoglobin A, hemoglobin A2, hemoglobin B, and hemoglobin F) are paralogs of each other. While each of these proteins serves the same basic function of oxygen transport, they have already diverged slightly in function: fetal hemoglobin (hemoglobin F) has a higher affinity for oxygen than adult hemoglobin. Function is not always conserved, however. Human angiogenin diverged from ribonuclease, for example, and while the two paralogs remain similar in tertiary structure, their functions within the cell are now quite different.
It is often asserted that orthologs are more functionally similar than paralogs of similar divergence, but several papers have challenged this notion.[32][33][34]
Paralogy regions
Sometimes, large chromosomal regions share gene content similar to other chromosomal regions within the same genome.[35] They are well characterised in the human genome, where they have been used as evidence to support the 2R hypothesis. Sets of duplicated, triplicated and quadruplicated genes, with the related genes on different chromosomes, are deduced to be remnants from genome or chromosomal duplications. A set of paralogy regions is together called a paralogon.[36] Well-studied sets of paralogy regions include regions of human chromosome 2, 7, 12 and 17 containing Hox gene clusters, collagen genes, keratin genes and other duplicated genes,[37] regions of human chromosomes 4, 5, 8 and 10 containing neuropeptide receptor genes, NK class homeobox genes and many more gene families,[38][39][40] and parts of human chromosomes 13, 4, 5 and X containing the ParaHox genes and their neighbors.[41] The Major histocompatibility complex (MHC) on human chromosome 6 has paralogy regions on chromosomes 1, 9 and 19.[42] Much of the human genome seems to be assignable to paralogy regions.[43]
Ohnology
Ohnologous genes are paralogous genes that have originated by a process of whole-genome duplication. The name was first given in honour of Susumu Ohno by Ken Wolfe.[44] Ohnologues are useful for evolutionary analysis because all ohnologues in a genome have been diverging for the same length of time (since their common origin in the whole genome duplication).
Xenology
Homologs resulting from horizontal gene transfer between two organisms are termed xenologs. Xenologs can have different functions, if the new environment is vastly different for the horizontally moving gene. In general, though, xenologs typically have similar function in both organisms. The term was coined by Walter Fitch.[15]
Gametology
Gametology denotes the relationship between homologous genes on non-recombining, opposite sex chromosomes. Gametologs result from the origination of genetic sex determination and barriers to recombination between sex chromosomes. Examples of gametologs include CHDW and CHDZ in birds.
Homology between sexes and forms
The term homology is sometimes applied to reproductive structures that share a common embryonic origin, but become spectacularly different between the two sexes in the adult. Those listed below are some of the more commonly cited examples.
| Male structure | Female structure | Notes |
|---|---|---|
| prostate | skene's gland | - |
| penis | clitoris | - |
| - | uterus | homologous to eggshell-depositing organs in reptiles and birds |
| bulbourethral gland | Bartholin's gland | - |
Among insects, the stinger used by infertile female worker bees is a modified ovipositor.
Homologies across phyla
See also
- Analogy (biology)
- Cladistics
- Deep homology
- OrthoDB
- Orthologous MAtrix (OMA)
- Protein family
- Protein superfamily
- TreeFam
- Syntelog
Notes
References
- ^ a b "Homology". Encyclopædia Britannica Online. Retrieved 2013-08-30.
- ^ a b Hyde, Janet Shibley; DeLamater, John D. (June 2010), "Chapter 5", Understanding Human Sexuality (11th ed.), New York: McGraw-Hill, p. 103, ISBN 978-0073382821
{{citation}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Bower, Frederick Orpen (1906). "Plant Morphology". Congress of Arts and Science: Universal Exposition, St. Louis, 1904. Houghton, Mifflin. p. 64.
- ^ Williams, David Malcolm; Forey, Peter L. (2004). Milestones in Systematics. CRC Press. p. 198. ISBN 0-415-28032-X.
- ^ "homogeneous, adj.". OED Online. March 2016. Oxford University Press. http://www.oed.com/view/Entry/88045? (accessed April 09, 2016).
- ^ "homogenous, adj.". OED Online. March 2016. Oxford University Press. http://www.oed.com/view/Entry/88055? (accessed April 09, 2016).
- ^ Scotland, R. W. (2010). "Deep homology: A view from systematics". BioEssays. 32 (5): 438–449. doi:10.1002/bies.200900175. PMID 20394064.
- ^ Cf. Butler, A. B.: Homology and Homoplasty. In: Squire, Larry R. (Ed.): Encyclopedia of Neuroscience, Academic Press, 2009, pp. 1195–1199.
- ^ Sattler R (1984). "Homology — a continuing challenge". Systematic Botany. 9 (4): 382–94. doi:10.2307/2418787. JSTOR 2418787.
- ^ Sattler, R. (1994). "Homology, homeosis, and process morphology in plants". In Hall, Brian Keith (ed.). Homology: the hierarchical basis of comparative biology. Academic Press. pp. 423–75. ISBN 0-12-319583-7.
- ^ Pinna, M. C. C. (1991). "Concepts and Tests of Homology in the Cladistic Paradigm". Cladistics. 7 (4): 367–394. doi:10.1111/j.1096-0031.1991.tb00045.x.
- ^ Brusca, R.C. & Brusca, G.J. 1990. Invertebrates. Sinauer Associates, Sunderland.: [i]-xviii, 1-922., P. 669
- ^ "Clustal FAQ #Symbols". Clustal. Retrieved 8 December 2014.
- ^ Koonin EV (2005). "Orthologs, paralogs, and evolutionary genomics". Annual Review of Genetics. 39: 309–38. doi:10.1146/annurev.genet.39.073003.114725. PMID 16285863.
- ^ a b Fitch WM (June 1970). "Distinguishing homologous from analogous proteins". Systematic Zoology. 19 (2): 99–113. doi:10.2307/2412448. PMID 5449325.
- ^ Falciatore A, Merendino L, Barneche F, et al. (January 2005). "The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas". Genes & Development. 19 (1): 176–87. doi:10.1101/gad.321305. PMC 540235. PMID 15630026.
- ^ Fang G, Bhardwaj N, Robilotto R, Gerstein MB (March 2010). "Getting started in gene orthology and functional analysis". PLoS Computational Biology. 6 (3): e1000703. doi:10.1371/journal.pcbi.1000703. PMC 2845645. PMID 20361041.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ COGs: Clusters of Orthologous Groups of proteins
Tatusov RL, Koonin EV, Lipman DJ (October 1997). "A genomic perspective on protein families". Science. 278 (5338): 631–7. doi:10.1126/science.278.5338.631. PMID 9381173. - ^ eggNOG: evolutionary genealogy of genes: Non-supervised Orthologous Groups
Muller J, Szklarczyk D, Julien P, et al. (January 2010). "eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations". Nucleic Acids Research. 38 (Database issue): D190–5. doi:10.1093/nar/gkp951. PMC 2808932. PMID 19900971. - ^ Inparanoid: Eukaryotic Ortholog Groups
Ostlund G, Schmitt T, Forslund K, et al. (January 2010). "InParanoid 7: new algorithms and tools for eukaryotic orthology analysis". Nucleic Acids Research. 38 (Database issue): D196–203. doi:10.1093/nar/gkp931. PMC 2808972. PMID 19892828. - ^ OrthoDB: The Hierarchical Catalog of Eukaryotic Orthologs
Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV (January 2011). "OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011". Nucleic Acids Research. 39 (Database issue): D283–8. doi:10.1093/nar/gkq930. PMC 3013786. PMID 20972218. - ^ OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes
Chen F, Mackey AJ, Stoeckert CJ, Roos DS (January 2006). "OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups". Nucleic Acids Research. 34 (Database issue): D363–8. doi:10.1093/nar/gkj123. PMC 1347485. PMID 16381887. - ^ Roundup
Deluca TF, Wu IH, Pu J, et al. (August 2006). "Roundup: a multi-genome repository of orthologs and evolutionary distances". Bioinformatics (Oxford, England). 22 (16): 2044–6. doi:10.1093/bioinformatics/btl286. PMID 16777906. - ^ OrthoMaM
Ranwez V, Delsuc F, Ranwez S, Belkhir K, Tilak MK, Douzery EJ (2007). "OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics". BMC Evolutionary Biology. 7: 241. doi:10.1186/1471-2148-7-241. PMC 2249597. PMID 18053139.{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ OrthologID
Chiu JC, Lee EK, Egan MG, Sarkar IN, Coruzzi GM, DeSalle R (March 2006). "OrthologID: automation of genome-scale ortholog identification within a parsimony framework". Bioinformatics. 22 (6): 699–707. doi:10.1093/bioinformatics/btk040. PMID 16410324. - ^ GreenPhylDB
Conte MG, Gaillard S, Lanau N, Rouard M, Périn C (January 2008). "GreenPhylDB: a database for plant comparative genomics". Nucleic Acids Research. 36 (Database issue): D991–8. doi:10.1093/nar/gkm934. PMC 2238940. PMID 17986457. - ^ TreeFam: Tree families database
Ruan J, Li H, Chen Z, et al. (January 2008). "TreeFam: 2008 Update". Nucleic Acids Research. 36 (Database issue): D735–40. doi:10.1093/nar/gkm1005. PMC 2238856. PMID 18056084. - ^ TreeFam: Tree families database
van der Heijden RT, Snel B, van Noort V, Huynen MA (2007). "Orthology prediction at scalable resolution by phylogenetic tree analysis". BMC Bioinformatics. 8: 83. doi:10.1186/1471-2105-8-83. PMC 1838432. PMID 17346331.{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Fulton DL, Li YY, Laird MR, Horsman BG, Roche FM, Brinkman FS (2006). "Improving the specificity of high-throughput ortholog prediction". BMC Bioinformatics. 7: 270. doi:10.1186/1471-2105-7-270. PMC 1524997. PMID 16729895.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E (February 2009). "EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates". Genome Research. 19 (2): 327–35. doi:10.1101/gr.073585.107. PMC 2652215. PMID 19029536.
- ^ Sayers EW, Barrett T, Benson DA, et al. (January 2011). "Database resources of the National Center for Biotechnology Information". Nucleic Acids Research. 39 (Database issue): D38–51. doi:10.1093/nar/gkq1172. PMC 3013733. PMID 21097890.
- ^ Studer RA, Robinson-Rechavi M (May 2009). "How confident can we be that orthologs are similar, but paralogs differ?". Trends in Genetics. 25 (5): 210–6. doi:10.1016/j.tig.2009.03.004. PMID 19368988.
- ^ Nehrt NL, Clark WT, Radivojac P, Hahn MW (June 2011). "Testing the ortholog conjecture with comparative functional genomic data from mammals". PLoS Computational Biology. 7 (6): e1002073. doi:10.1371/journal.pcbi.1002073. PMC 3111532. PMID 21695233.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Eisen, Jonathan. "Special Guest Post & Discussion Invitation from Matthew Hahn on Ortholog Conjecture Paper".
- ^ Lundin, LG (April 1993). "Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse". Genomics. 16 (1): 1–19. doi:10.1006/geno.1993.1133. PMID 8486346.
- ^ Coulier, F; Popovici, C; Villet, R; Birnbaum, D (Dec 15, 2000). "MetaHox gene clusters". The Journal of experimental zoology. 288 (4): 345–51. doi:10.1002/1097-010X(20001215)288:4<345::AID-JEZ7>3.0.CO;2-Y. PMID 11144283.
- ^ Ruddle, FH; Bentley, KL; Murtha, MT; Risch, N (1994). "Gene loss and gain in the evolution of the vertebrates". Development (Cambridge, England). Supplement: 155–61. PMID 7579516.
- ^ Pébusque, MJ; Coulier, F; Birnbaum, D; Pontarotti, P (September 1998). "Ancient large-scale genome duplications: phylogenetic and linkage analyses shed light on chordate genome evolution". Molecular Biology and Evolution. 15 (9): 1145–59. doi:10.1093/oxfordjournals.molbev.a026022. PMID 9729879.
- ^ Larsson, TA; Olsson, F; Sundstrom, G; Lundin, LG; Brenner, S; Venkatesh, B; Larhammar, D (Jun 25, 2008). "Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions". BMC Evolutionary Biology. 8: 184. doi:10.1186/1471-2148-8-184. PMC 2453138. PMID 18578868.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Pollard, Sophie L.; Holland, Peter W.H. (2000). "Evidence for 14 homeobox gene clusters in human genome ancestry". Current Biology. 10 (17): 1059–1062. doi:10.1016/S0960-9822(00)00676-X.
- ^ Mulley, JF; Chiu, CH; Holland, PW (Jul 5, 2006). "Breakup of a homeobox cluster after genome duplication in teleosts". Proceedings of the National Academy of Sciences of the United States of America. 103 (27): 10369–72. doi:10.1073/pnas.0600341103. PMC 1502464. PMID 16801555.
- ^ Flajnik, Martin F.; Kasahara, Masanori (2001). "Comparative Genomics of the MHC". Immunity. 15 (3): 351–362. doi:10.1016/S1074-7613(01)00198-4.
- ^ McLysaght, Aoife; Hokamp, Karsten; Wolfe, Kenneth H. (2002). "Extensive genomic duplication during early chordate evolution". Nature Genetics. 31 (2): 200–204. doi:10.1038/ng884.
- ^ Wolfe K (May 2000). "Robustness--it's not where you think it is". Nature Genetics. 25 (1): 3–4. doi:10.1038/75560. PMID 10802639.
Further reading
- Carroll, Sean B. (2006). Endless Forms Most Beautiful. New York: W.W. Norton & Co. ISBN 0-297-85094-6.
- Carroll, Sean B. (2006). The making of the fittest: DNA and the ultimate forensic record of evolution. New York: W.W. Norton & Co. ISBN 0-393-06163-9.
- DePinna MC (1991). "Concepts and tests of homology in the cladistic paradigm". Cladistics. 7 (4): 367–94. doi:10.1111/j.1096-0031.1991.tb00045.x.
- Dewey CN, Pachter L (April 2006). "Evolution at the nucleotide level: the problem of multiple whole-genome alignment". Human Molecular Genetics. 15 (Spec No 1): R51–6. doi:10.1093/hmg/ddl056. PMID 16651369.
- Fitch WM (May 2000). "Homology a personal view on some of the problems". Trends in Genetics. 16 (5): 227–31. doi:10.1016/S0168-9525(00)02005-9. PMID 10782117.
- Gegenbaur, G. (1898). Vergleichende Anatomie der Wirbelthiere... Leipzig.
{{cite book}}: CS1 maint: location missing publisher (link) - Haeckel, Е. (1866). Generelle Morphologie der Organismen. Bd 1-2. Вerlin.
{{cite book}}: CS1 maint: location (link) CS1 maint: location missing publisher (link) - Owen, R. (1847). On the archetype and homologies of the vertebrate skeleton. London.
- Mindell DP, Meyer A (2001). "Homology evolving" (PDF). Trends in Ecology and Evolution. 16 (8): 434–40. doi:10.1016/S0169-5347(01)02206-6.
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA (November 2008). "The quest for orthologs: finding the corresponding gene across genomes". Trends Genet. 24 (11): 539–51. doi:10.1016/j.tig.2008.08.009. PMID 18819722.
External links
- Brigandt, Ingo (2011) "Essay: Homology." In: The Embryo Project Encyclopedia. ISSN 1940-5030. http://embryo.asu.edu/handle/10776/1754

