User:Aaadddaaammm/Chlamy
![]() There is already an article named Chlamydomonas reinhardtii in the mainspace.
There is already an article named Chlamydomonas reinhardtii in the mainspace.
| Aaadddaaammm/Chlamy | |
|---|---|
| |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | c. reinhardtii
|
| Binomial name | |
| Chlamydomonas reinhardtii | |
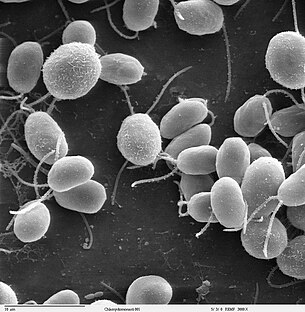
Chlamydomonas reinhardtii is a haploid single celled green alga about 10 micrometres in diameter. C. reinhardtii is widely distributed worldwide in soil and fresh water, however it is primarily used as a model organism in biology.
Swims with two flagella. They have a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an "eyespot" that senses light.
Although in a wide range of subfields. When illuminated, C. reinhardtii can grow in medium lacking carbon and energy sources, and can also grow in the dark when supplied with these. C. reinhardtii is also of interest in the biofuel field, as a source of hydrogen.
History[edit]
The C. reinhardtii wild type laboratory strain c137 (mt+) originates from an isolate made near Amherst, Massachusetts, in 1945 by Gilbert M. Smith.[1][2] The lineage of widely used laboratory strains has been well reviewed by XXX (refXXX).
The species has been spelled several different ways because of different transliterations of the name from Russian: reinhardi, reinhardii and reinhardtii all refer to the same species, C. reinhardtii Dangeard.[3]
Model organism[edit]
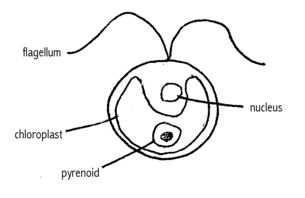
C. reinhardtii is the most commonly used unicellular photosynthetic eukaryote in biological research. Its simple structure gives it many advantages over other model organisms in a laboratory. Photosynthesis and chloroplast research is much simpler in C. reinhardtii over the leading model plant, Arabidopsis thaliana, as C. reinhardtii lacks the complicated tissue structure of a higher plant and has a much faster generation time. The presence of flagella in C. reinhardtii gives it use compared to other unicellular eukaryotes, such as yeast. In addition, C. reinhardtii is often used as a model alga, often in the field of biofuels. Simple genetics and ease of growth in the lab has also lead to the emergence of C. reinhardtii as a leading model organisms in many other fields of reasearch, such as proteomics and circadian rhythms.
A number of resources are available for C. reinhardtii. In 2007, its complete nuclear genome sequence was published[4], which was added to the previously sequenced plastome and mitochondriome. From these sequences a number of microarrays have been developed refXXX. Tranformation of these 3 genomes is routine, as are other mutation protocols, which has lead to the accumulation of a large mutant library for C. reinhardtii.
Channelrhodopsin-1 and Channelrhodopsin-2, proteins that function as light-gated cation channels, were originally isolated from C. reinhardtii.[5][6]. These proteins and others like them are increasingly widely used in the field of optogenetics.[citation needed]
Reproduction[edit]
Vegetative cells of C. reinhardtii are haploid with 17 small chromosomes. Under nitrogen starvation, haploid gamete develop. There are two mating type, identical in appearance and known as mt(+) and mt(-), which can fuse to form a diploid zygote. The zygote is not flagellated, and it serves as a dormant form of the species in the soil. When exposed to light the zygote undergoes meiosis and releases four flagellated haploid cells that resume the vegetative life cycle.
Under ideal growth conditions, cells may sometimes undergo two or three rounds of mitosis before the daughter cells are released from the old cell wall into the Growth medium. Thus, a single growth step may result in 4 or 8 daughter cells per mother cell.
The cell cycle of this unicellular green algae can be synchronized by alternating periods of light and dark. The growth phase is dependent on light, whereas, after a point designated as the transition or commitment point, processes are light-independent.[7]
Genetics[edit]
The attractiveness of the alga as a model organism has increased with the release of several genomic resources to the public domain. The current draft of the C. reinhardtii nuclear genome sequence prepared by Joint Genome Institute of the U.S. Dept of Energy comprises 78 scaffolds totaling approximately 112 Megabase. The current assembly of the nuclear genome is available online.[8]
The ~15.8 Kilobase mitochondrial genome is available online at the NCBI database [9], as is the more recently completed >200 Kb chloroplast genome [10].
In addition to genomic sequence data there is a large supply of expression sequence data available as cDNA libraries and expressed sequence tags (ESTs). Over 30 cDNA libraries are currently available online[11], and there are more than 200,000 EST sequences available[12]. A BAC library can be purchased from the Clemson University Genomics Institute[13].
Evolution[edit]
Chlamydomonas has been used to study different aspects of evolutionary biology and ecology. It is an organism of choice for many selection experiments because (1) it has a short generation time, (2) it is both a heterotroph and facultative autotroph, (3) it can reproduce both sexually and asexually, and (4) there is a wealth of genetic information already available.
Some examples of evolutionary work done with Chlamydomonas include the evolution of sexual reproduction,[14] the fitness effect of mutations,[15] and the effect of adaptation to different levels of CO2.[16]
DNA transformation techniques[edit]
Gene transformation occurs mainly by homologous recombination in the chloroplast and heterologous recombination in the nucleus. The C. reinhardtii chloroplast genome can be transformed using microprojectile particle bombardment and the nuclear genome has been transformed with both glass bead agitation and electroporation. The microprojectile particle bombardment procedure appears to be the most efficient way of introducing DNA into the chloroplast genome. This is probably because the chloroplast occupies over half of the volume of the cell providing the microprojectile with a large target. Electroporation has been shown to be the most efficient way of introducing DNA into the nuclear genome with maximum transformation frequencies two orders of magnitude higher than obtained using glass bead method.[citation needed]
Hydrogen production[edit]
In 1939 the German researcher Hans Gaffron (1902–1979) discovered the hydrogen metabolism of unicellular green algae. C. reinhardtii and some other green algae can, under specified circumstances, stop producing photosynthetic oxygen and convert instead to the production of hydrogen. This reaction is catalysed by an oxygen-sensitive enzyme called hydrogenase. Over the next thirty years Gaffron and his team worked out the basic mechanics of this photosynthetic hydrogen production by algae.[17]
Notes[edit]
- ^ "CC-125 wild type mt+ 137c". Chlamydomonas Center core collection list.
- ^ The Chlamydononas Sourcebook, ISBN 978-0-12-370873-1)
- ^ http://megasun.bch.umontreal.ca/protists/chlamy/taxonomy.html Chlamydomonas Taxonomy.
- ^ Merchant; Prochnik, SE; Vallon, O; Harris, EH; Karpowicz, SJ; Witman, GB; Terry, A; Salamov, A; Fritz-Laylin, LK; et al. (2007). "The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions". Science. 318 (5848): 245–250. doi:10.1126/science.1143609. PMC 2875087. PMID 17932292.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Nagel G, Ollig D, Fuhrmann M; et al. (June 28, 2002). "Channelrhodopsin-1: a light-gated proton channel in green algae". S cience. 296 (5577): 2395–8. doi:10.1126/science.1072068. PMID 12089443.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Lagali PS, Balya D, Awatramani GB, Münch TA, Kim DS, Busskamp V, Cepko CL, Roska B (June 2008). "Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration". Nature Neuroscience. 11 (6): 667–75. doi:10.1038/nn.2117. PMID 18432197.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Oldenhof, H, Zachleder, V. and van den Ende, H. 2006. Blue- and red-light regulation of the cell cycle in Chlamydomonas reinhardtii (Chlorophyta). Eur. J. Phycol. 41: 313 - 320
- ^ http://www.phytozome.net/chlamy.php
- ^ http://www.ncbi.nlm.nih.gov/nuccore/NC_001638
- ^ http://www.ncbi.nlm.nih.gov/nuccore/NC_005353
- ^ http://www.ncbi.nlm.nih.gov/UniGene/lbrowse2.cgi?TAXID=3055
- ^ http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=3055&lvl=3&lin=f&keep=1&srchmode=1&unlock
- ^ http://jupiter.biology.duke.edu/libraries.html
- ^ Colegrave N. 2002. Sex releases the speed limit on evolution. Nature 420: 664-666.
- ^ De Visser et al. 1996 The effect of sex and deleterious mutations on fitness in Chlamydomonas. Proc. R. Soc. Lond. B 263-193-200.
- ^ Collins & Bell. 2004. Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga. Nature 431: 566-569.
- ^ Anastasios Melis, Thomas Happe (2004). "Trails of green alga hydrogen research — from Hans Gaffron to new frontiers" (PDF). Photosynthesis Research. 80 (1–3): 401–409. doi:10.1023/B:PRES.0000030421.31730.cb. PMID 16328836.
Further reading[edit]
Aoyama, H., Kuroiwa, T. and Nakamura, S. 2009. The dynamic behaviour of mitochondria in living zygotes during maturation and meiosis in Chlamydomonas reinhardtii. Eur. J. Phycol. 44: 497 - 507.
Jamers, A., Lenjou, M., Deraedt, P., van Bockstaele, D., Blust, R. and de Coen, W. 2009. Flow cytometric analysis of the cadmium-exposed green algae Chlamydomonas reinhadtii (Chlorophyceae). Eur. J. Phcol. 44: 54 - 550.
External links[edit]
- Chlamydomonas reinhardtii resources at the Joint Genome Institute
- The Chlamydomonas Center - genomic, genetic and bibliographic information and the Chlamydomonas culture collection.
- Chlamydomonas reinhardtii cell, life cycle, strains, mating types
- "Aaadddaaammm/Chlamy" at the Encyclopedia of Life
- Guiry, M.D.; Guiry, G.M. "'Chlamydomonas reinhardtii'". AlgaeBase. World-wide electronic publication, National University of Ireland, Galway.
Category:Green algae
Category:Model organisms
Category:Hydrogen production
Category:Sequenced genomes
