User:Dolleyj/sandbox
Introduction
[edit]Pharmacogenomics (a combination of pharmacology and genomics) is the study of how genetic makeup influences an individual’s response to drugs. [1] It deals with the influence of genetic variation on drug response in patients by correlating gene expression or single-nucleotide polymorphisms with a drug's efficacy or toxicity. [2] By doing so, pharmacogenomics aims to develop rational means to optimize drug therapy, with respect to the patients' genotype, to ensure maximum efficacy with minimal adverse effects. [3] Such approaches promise the advent of "personalized medicine"; in which drugs and drug combinations are optimized, or even avoided, based on each individual's unique genetic makeup. [4][5] Since the advent of medicine, itself, it has been well-established that no two individuals respond exactly the same to any given drug. Also, it is known that while a drug may work well in the vast amount of a population, it may be ineffective, or even harmful, to a select subset of the population. Pharmacogenomics seeks to explain the mechanisms behind these occurrences and provide better and safer patient care. In order to provide pharmacogenomic based recommendations for a given drug, several possible types of input can be used: genotyping, exome sequencing, or wholegenome sequencing. [6]
Pharmacodynamic Genes
[edit]A pharmacodynamic gene is the gene that encodes the protein which is the target of a drug [7]. These genes can code for cell surface receptors, enzymes, or nuclear hormone receptors. Drugs directly interact with these proteins to produce the drug’s therapeutic effects. These interactions divide drugs into two basic mechanisms of action: agonist or antagonist.
An agonist drug binds to the target protein, and activates a cellular pathway to produce the desired therapeutic effect.
An antagonist drug binds to the target protein, and inhibits the binding of that protein’s activator. This prevents the activation of a cellular pathway to produce the desired therapeutic effect.
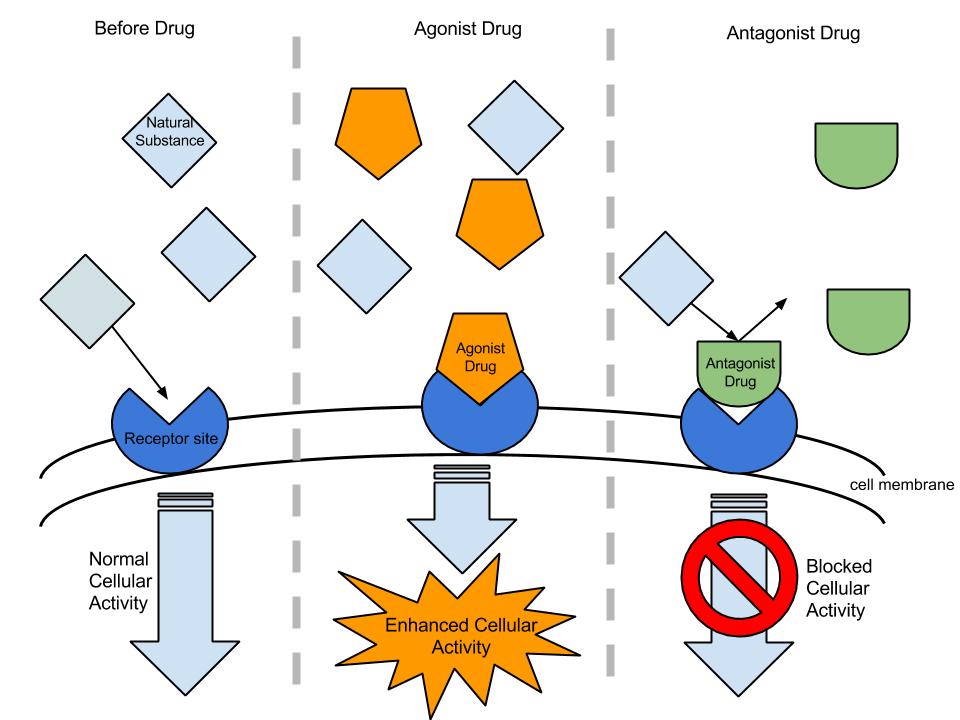
Examples of Agonist & Antagonist Drugs
[edit]An example of an agonist drug would be the dopamine agonists drugs (bromocriptine, pergolide, pramipexole, ropinirole, piribedil, cabergoline, apomorphine, and lisuride) used to treat Parkinson’s Disease patients. These drugs bind to the dopaminergic postsynaptic receptors in the brain to activate the signaling pathways necessary for gene transcription and production of dopamine.
An example of an antagonist drug would be trastuzumab (brand name Herceptin). Trastuzumab targets the receptor tyrosine-protein kinase ERBB-2. ERBB-2, also called HER2 (from its name human epidermal growth factor receptor 2), promotes cell growth and division under normal conditions; however, when ERBB-2 is expressed, it is carcinogenic and found in 20% of early stage breast cancers [7]. When administered, trastuzumab binds to the ERBB-2 receptor, thus inhibiting carcinogenesis.
Genetic variation in pharmacodynamic genes
[edit]Single nucleotide polymorphisms (SNPs) are genetic variations where a single nucleotide is found to persist in different allelic forms across members of a human population[7]. While not all SNPs may affect genes, or downstream proteins, many result in variation of pharmacodynamic genes that greatly impact the structure or chemistry of the drug targeted proteins. In turn, this affects the interaction of the drug with its target protein, and result in the drug exhibiting an undesired effect.
Example
[edit]Warfarin (Brand name: Coumadin) is an anticoagulant drug prescribed to patients with prosthetic heart valves or atrial fibrillation to prevent systemic embolism. Warfarin’s mechanism of action is to act as a vitamin K antagonist. Warfarin enters the body as a prodrug; therefore it must be first metabolized before it is able to produce its therapeutic effect. It is oxidized by the CYP2C9 enzyme in the liver becoming active. Then it inhibits the vitamin K epoxide reductase complex, subunit 1 (VKORC1) to exhibit its anticoagulant effect. While an effective drug, warfarin unfortunately has a narrow window for maintaining a therapeutic INR, which is between 2.0 to 3.0. An INR greater than 4.0 is supratherapeutic and greatly increases the risk for bleeding. Because of this narrow therapeutic window, it takes time to achieve a stable therapy [8].
Three SNPs have been discovered to impact the metabolism and effectiveness of warfarin. Identification of these SNPs in carriers greatly influences the dosage for those individuals and aids in safer start dosages when beginning a regular warfarin therapy regimen. Two SNPs are located in the CYP2P9 gene and one SNP is in the VKORC1 gene. For purposes of the article, VKORC1 SNP will be detailed, as VKORC1 is the primary pharmacodynamic gene of warfarin.
At the 1639 SNP loci in the VKORC1 gene, the common allele, G, is replaced by the A allele, resulting in a decreased production of the VKORC1 protein. A dose of warfarin (considered normal or typical for a G allele person) in the presence of a decreased concentration of VKORC1 protein would result in an increased therapeutic effect (increased anticoagulation). Therefore, people carrying an A allele need a lower warfarin dose to produce appropriate anticoagulation. The prevalence of the A allele is found in 37% of Caucasians and 14% of Africans[8].
Clinically, this sensitivity to warfarin translates into individualized dosages for aberrant SNP carriers. People carrying an A allele generally require 28% less warfarin dose per allele than non-A allele carriers. Similar dosage adjustments are made for carriers of the CYP2C9*2 and CYP2C9*3 alleles, 19% and 33% dose reduction, respectively[8]. The table below illustrates the recommended dosages for carriers of the warfarin SNP alleles.
Pharmacokinetic Genes
[edit]A pharmacokinetic gene is a gene that codes for proteins that are involved in the absorption, distribution, metabolism, or excretion (ADME) of a drug.[9] Pharmacokinetic genes encode a diverse array of proteins such as transporters, ion channels, cytochrome P450, and structural proteins. Pharmacokinetic genes can alter the bioavailability of a drug by regulating the concentration of drug in the blood and tissues.
The Effect of Pharmacokinetic Genes on ADME
[edit]- Absorption: The route of administration determines the availability of the drug within the body. Absorption usually occurs within the gastrointestinal tract for drugs that are ingested and within the lungs for drugs that are administered via inhalation. Using an intravenous route of administration results in no absorption of the drug because the drug is injected directly into the blood stream.[10] Variation in absorption-based pharmacokinetic genes can affect gastrointestinal cell transporter and channel structure and activity, which can affect the absorption of a drug.
- Distribution: The action of the cardiovascular system is the primary mechanism by which a drug is distributed throughout the body. Distribution of the drug may face difficulty at specific sites within the body such as the hematoencephalic barrier.[11] Variation in distribution-based pharmacokinetic genes can alter the integrity of the hematoencephalic barrier allowing more or less distribution to the brain.
- Metabolism: The majority of drugs are enzymatically broken down by the action of the hepatic cytochrome P450 family of redox enzymes. Cytochrome P450 enzyme activity on drug substrate can change the pharmacology of a drug. Some of the changes that can occur are activation of a prodrug or production of a pharmacologically inert metabolite.[12] Variation in metabolism-based pharmacokinetic genes can alter the activity of cytochrome P450 enzymes, which will affect the pharmacology of a drug, the rate of elimination amongst other things.
- Excretion (Elimination) : The body rids of the drug primarily through the mechanism of excretion in the urine and/or in solid waste. Variation in elimination-based pharmacokinetic genes can affect the activity of the nephron and nephrological proteins that pertain to the excretion of drug metabolites in the urine.
Genetic Variation in Pharmacokinetic Genes
[edit]Variation in the nucleotide sequence of a pharmacokinetic gene can alter the structure and chemistry of the encoded protein product. Single nucleotide polymorphisms may or may not have an affect on pharmacokinetic genes depending on the position of the mutation in the gene. In addition to mutations in a pharmacokinetic gene itself, nucleotide variation in non-protein coding regulatory regions can also affect the expression of a pharmacokinetic gene. [13]
Atorvastatin (Lipitor) is a statin-based small molecule drug that is administered to lower cholesterol levels. Atorvastatin targets mainly the liver and hepatic enzymes. It is a competitive inhibitor of hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductase. In the mevalonate pathway, HMG-CoA reductase functions as the rate-determining enzyme for cholesterol production as it catalyzes the reduction of HMG-CoA to mevalonate.[14]
Three single-nucleotide polymorphisms have been associated with affecting Atorvastatin activity. These SNPs are associated with the HMG-CoA gene (HMGCR), the kinesin-like protein (KIF6), and the multidrug resistance protein-1 (ABCB1). At the 14863 SNP loci of the HMGCR gene the common allele, A, is changed to the mutant allele T. This SNP in the HMGCR gene will cause a decrease in HMG-CoA's response to statin drugs such as Atorvastatin.[15] At the 2250 SNP loci of the KIF6 gene a missense mutation is occurring such that the common allele, T, is changed to the mutant allele, C. This SNP in the KIF6 gene will cause an increased response of kinesin-like protein to Atorvastatin.[16] At the 208920 SNP loci of the ABCB1 gene, the common allele, A, is changed to the mutant allele, T. This SNP in the ABCB1 gene causes a large decrease in low-density lipoprotein and a slight increase in high-density lipoprotein cholesterol in females.[17]
Controversies
[edit]Some alleles that vary in frequency between specific populations have been shown to be associated with differential responses to specific drugs. The beta blocker Atenolol is an anti-hypertensive medication that is shown to more significantly lower the blood pressure of Caucasian patients than African American patients in the United States. This observation suggests that Caucasian and African American populations have different alleles governing oleic acid biochemistry, which react differentially with Atenolol.[18] Similarly, hypersensitivity to the antiretroviral drug abacavir is strongly associated with a single-nucleotide polymorphism that varies in frequency between populations.[19]
This brings up the concept of race-based medicine, which is a very controversial subject. [20] . It is a scientific fact that similar population such as ethnicities or races have similar genetics, thus, similar races may have similar patterns of SNPs. A drug's effectiveness is dependent upon an individual's genome and any SNPs that lie within the genes or in the regulatory genes that are associated with a drug's response. Therefore, it seems logical to develop and prescribe specific drugs to certain populations due solely to similarities in their genomic makeup; yet, society has condemned this idea.
For example, the FDA approval of the drug BiDil with a label specifying African-Americans with congestive heart failure, produced a storm of controversy over race-based medicine and fears of genetic stereotyping, even though the label for BiDil did not specify any genetic variants but was based on racial self-identification. [21]
Applications
[edit]Several gene chips and systems for genotyping genes associated with drug metabolism have been developed. One of the chips, the first approved by the FDA, is the AmpliChip, which types alleles of the CYP2C19 and CYP2D6 genes, whose mutations are associated with decreased or increased drug metabolism. The AmpliChip, a product from Roche, was the first approved diagnostic genotyping product by the FDA. The product was approved in 2005 and can identifys up to 33 CYP2D6 alleles and 3 CYP2C19 alleles. It is a microarray and uses Polymerase chain reaction amplification of DNA isolated from the patient. The PCR products are applied to the AmpliChip. The chip contains oligonucleotide probes that hybridize with the PCR products. The probes represent the different polymorphism of the gene. In a clinical setting the information from the gene can be used as a component in determining prescribed drug dosage.[22]
The ability of the gene chips to identify the degree of metabolism can be determined with the use of a probed drug. The probe drug is administered to the patient and then the drug and its resulting metabolites are measured from the patient’s urine. From this test a, a patient can be determined to be an, extensive (EM), poor (PM), ultra rapid (UM) or intermediate (IM) metabolizers. To determines these values the, ratio of dextromethorphan, the probe drug, to dextrorphan , the metabolite, can be determined from the urine. When the genotypes of samples obtained from PCR methods is compared with the genotype indicated by the AmpliChip the correspondence is nearly 100%. In determining whether a patient has an EM, UM, IM or Em phenotype, the amplichip was able to identify correctly 80% of samples. To separate the patients into phenotypes determined from the genotypes, the alleles reported by the AmpliChip were used. If a patient had one non-functional allele, one reduced activity allele or two reduced activity alleles, they were characterized as IM and EM, if they had at least one function allele, and UM if they had at least three copies of a functional allele.
Comparing the predicted phenotype from the chip with probe drug data, 15 out of 15 were correctly predicted. For IM, 8 out of 19 were correctly predicted, for EM 106 out of 111 were correctly identified and for UM only 1 out of 17 were correctly identified. The possible reasons for the poor result for UMmay be that factors independent of theCYP2D6 may be at play, such as other genes.
Other systems and gene chips on the market include the INFINITI 2C9 & VKORC1 Multiplex Assay for Warfarin, Tag-it mutation Detection System, and Invader UGT1A1 Molecular Assay. In tests to determine the accuracy of these systems, 112 subjects were genotyped. The Infinite system was found to be a 100% accurate for CYP2C9 *2 and *3 and VKORC1-1639 SNPs. the invader was also 100% accurate. The tag it mutation system was also 100% for CYP2C9*2 and *3 and 99% accurate for VKORCI genotyping.
AN important factor besides accuracy is turnaround time for a test, especially in a hospital setting. The three systems mentioned above were tested for turnaround time, the amount of time from the beginning of the test to results. The invader system had the lowest turnaround time at about 3 hours. The Infiniti and tag it systems both had system had a turnaround of 8 hours. Another important parameter is labor time. The invader system used 1.42 hours of labor time. To determine dose, dosing algoritms can be used, which incorporate clinical information and genotype information. A number of patients on a stable dose of warfarin were used as the subjects in a test to determine if dosing algorithms which incorporate genotypes were accurate. These algorithms incorporated the CYP2C9 and VKORC1 variants.[23]
The average age of a patient was 67 years and 62% were male, 57% were Caucasian, 19% African American, Hispanic 14 % and 10% Asian. The 12 algorithms that incorporated genotypes explained between 33% and 55% of the variation in warfarin dosage[24]
| Product | Company | Submission |
|---|---|---|
| xTAG CYP2D6 Kit v3 | Luminex Molecular Diagnostics, Inc. | K130189, K093420 |
| Spartan RX CYP2C19 Test | System Spartan Bioscience, Inc. | K123891 |
| Verigene CYP2C 19 Nucleic Acid Test | Nanosphere, Inc. | K120466 |
| INFINITI CYP2C19 Assay | AutoGenomics, Inc. | K101683 |
| Invader UGT1A1 Molecular Assay | Third Wave Technologies Inc. | K051824 |
| Roche AmpliChip CYP450 microarray | Roche Molecular Systems, Inc. | K043576, K042259 |
| eSensor Warfarin Sensitivity Saliva Test | GenMark Diagnostics | K110786 |
| eQ-PCR LC Warfarin Genotyping kit | TrimGen Corporation | K073071 |
| eSensor Warfarin Sensitivity Test and XT-8 Instrument | Osmetech Molecular Diagnostics | K073720 |
| Gentris Rapid Genotyping Assay - CYP2C9 & VKORCI | ParagonDx, LLC | K071867 |
| INFINITI 2C9 & VKORC1 Multiplex Assay for Warfarin | AutoGenomics, Inc. | K073014 |
| Verigene Warfarin Metabolism Nucleic Acid Test and Verigene System | Nanosphere, | Inc. K070804 |
Future Directions
[edit]In the future, high throughput tests genotypes of Metabolism enzymes and drug transporters that effect Absorption, distribution, metabolism and elimination (ADME) will be available. One such system, approved for clinical research, is the DMET system (Drug Metabolizing Enzyme and Transporters), which provides wide coverage of 1913 genetic variants, related to pharmacogenetics including drug transporters in cells and covers 225 genes. The system uses a molecular inversion probe, with the advantages that less and lower quality DNA can used. The results from DMET system was compared to lower through put genotyping systems. A total of 19,942 SNP patient pairs were tested, with DMET compared against at least one orthogonal method. The DMET system was able to identify genotypes with 99.9% accuracy.[25]
See also
[edit]References
[edit]- ^ Ermak G (February 2013). Modern Science & Future Medicine (second ed.). CreateSpace Independent Publishing Platform. pp. 164 year = 2013. ISBN 978-1-4823-0885-3.
{{cite book}}: Missing pipe in:|pages=(help) - ^ Wang L (2010). "Pharmacogenomics: a systems approach". Wiley Interdiscip Rev Syst Biol Med. 2 (1): 3–22. doi:10.1002/wsbm.42. PMC 3894835. PMID 20836007.
- ^ Becquemont L (June 2009). "Pharmacogenomics of adverse drug reactions: practical applications and perspectives". Pharmacogenomics. 10 (6): 961–9. doi:10.2217/pgs.09.37. PMID 19530963.
- ^ "Guidance for Industry Pharmacogenomic Data Submissions" (PDF). U.S. Food and Drug Administration. March 2005. Retrieved 2008-08-27.
- ^ Squassina A, Manchia M, Manolopoulos VG, Artac M, Lappa-Manakou C, Karkabouna S, Mitropoulos K, Del Zompo M, Patrinos GP (August 2010). "Realities and expectations of pharmacogenomics and personalized medicine: impact of translating genetic knowledge into clinical practice". Pharmacogenomics. 11 (8): 1149–67. doi:10.2217/pgs.10.97. PMID 20712531.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Huser, V.; Cimino, J. J. (2013). "Providing pharmacogenomics clinical decision support using whole genome sequencing data as input". AMIA Summits on Translational Science Proceedings AMIA Summit on Translational Science. 2013: 81. PMC 3814493. PMID 24303303.
- ^ a b c al.], Joel T. Dudley ... [et (2013). Exploring personal genomics. Oxford: Oxford University Press. pp. 139–159. ISBN 978-0-19-964448-3.
- ^ a b c "Warfarin Dosing and VKORC1/CYP2C9". Retrieved April 5, 2014.
- ^ Dudley, Joel (2013). Exploring personal genomics. Oxford: Oxford University Press. ISBN 9780199644483.
- ^ Thomas, Gareth (2007). Medicinal chemistry (2nd ed., Extensively rev. and updated. ed.). Chichester: John Wiley. ISBN 9780470025970.
- ^ Thomas, Gareth (2007). Medicinal chemistry (2nd ed., Extensively rev. and updated. ed.). Chichester: John Wiley. ISBN 9780470025970.
- ^ Thomas, Gareth (2007). Medicinal chemistry (2nd ed., Extensively rev. and updated. ed.). Chichester: John Wiley. ISBN 9780470025970.
- ^ Dudley, Joel (2013). Exploring personal genomics. Oxford: Oxford University Press. ISBN 9780199644483.
- ^ Trevor, edited by Bertram G. Katzung ; associate editors, Susan B. Masters, Anthony J. (2009). Basic & clinical pharmacology (11th ed.). New York: McGraw-Hill Medical. ISBN 9780071604055.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ Chasman, D (2004). "Pharmacogenetic study of statin therapy and cholesterol reduction". JAMA. 291 (23): 2821–2827. doi:10.1001/jama.291.23.2821. PMID 15199031. Retrieved 1 May 2014.
- ^ Iakoubova, C (2008). "Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials". Journal of the American College of Cardiology. 51 (4). J Am Coll Cardiol: 435–443. doi:10.1016/j.jacc.2007.05.057. PMID 18222353. Retrieved 1 May 2014.
- ^ Kajinami, K (2004). "Polymorphisms in the multidrug resistance-1 (MDR1) gene influence the response to atorvastatin treatment in a gender-specific manner". The American Journal of Cardiology. 93 (8). J Am Cardiol: 1046–1050. doi:10.1016/j.amjcard.2004.01.014. PMID 15081455. Retrieved 1 May 2014.
- ^ Wikoff, William (2013). "Pharmacometabolomics Reveals Racial Differences in Response to Atenolol Treatment". PLOS ONE. 8 (3): e57639. doi:10.1371/journal.pone.0057639. PMC 3594230. PMID 23536766.
- ^ Rotimi, Charles (2010). "Ancestry and Disease in the Age of Genomic Medicine". New England Journal of Medicine. 363 (16). NEJM: 1551–1558. doi:10.1056/NEJMra0911564. PMID 20942671. Retrieved 1 May 2014.
- ^ Krimsky, edited by Sheldon (2011). Race and the genetic revolution : science, myth, and culture. New York: Columbia University Press. ISBN 978-0231156974.
{{cite book}}:|first=has generic name (help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Krimsky, eSheldon (2011). Race and the genetic revolution : science, myth, and culture. New York: Columbia University Press. ISBN 978-0231156974.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Rebsamen MC, Desmeules J, Daali Y, et al. The AmpliChip CYP450 test: cytochrome P450 2D6 genotype assessment and phenotype prediction. Pharmacogenomics J. 2009;9(1):34-41.
- ^ Langley MR, Booker JK, Evans JP, Mcleod HL, Weck KE. Validation of clinical testing for warfarin sensitivity: comparison of CYP2C9-VKORC1 genotyping assays and warfarin-dosing algorithms. J Mol Diagn. 2009;11(3):216-25.
- ^ Lubitz SA, Scott SA, Rothlauf EB, et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J Thromb Haemost. 2010;8(5):1018-26.
- ^ Fernandez CA, Smith C, Yang W, et al. Concordance of DMET plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther. 2012;92(3):360-5.
Further reading
[edit]- Katsnelson A (August 2005). "A Drug to Call One's Own: Will medicine finally get personal?". Scientific American.
- Karczewski KJ, Daneshjou R, Altman RB (2012). "Chapter 7: Pharmacogenomics". PLOS Comput. Biol. 8 (12): e1002817. doi:10.1371/journal.pcbi.1002817. PMC 3531317. PMID 23300409.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
External links
[edit]- "Pharmacogenomics Factsheet". National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine. Retrieved 2011-07-11.
a quick introduction to customised drugs
- "Pharmacogenomics Education Initiatives". U.S. Food and Drug Administration. 2010-09-24. Retrieved 2011-07-11.
- "Table of Pharmacogenomic Biomarkers in Drug Labels". U.S. Food and Drug Administration. 2011-05-23. Retrieved 2011-07-11.
- "The Pharmacogenomics Knowledge Base". PharmGKB. Stanford University. Retrieved 2011-07-11.
- "Personalized Medicine (Pharmacogenetics)". University of Utah's Genetic Science Learning Center. Retrieved 2011-07-11.
Journals:
- "Future Medicine - Pharmacogenomics". Journal. Future Medicine Ltd. ISSN 1462-2416.
- "Pharmacogenetics and Genomics". Journal (previously Pharmacogenetics). Lippincott Williams & Wilkins. ISSN 1744-6872. Retrieved 2011-07-11.
- "The Pharmacogenomics Journal". Nature Publishing Group. ISSN 1470-269X. Retrieved 2011-07-11.
- "Pharmacogenomics : Subjects : Omics Gateway". Nature Publishing Group. Retrieved 2011-07-11.

