Glossary of cellular and molecular biology (0–L): Difference between revisions
Content deleted Content added
m →References: Page is over [[WP:PEIS] limit; use <references /> tag directly so references render as far as possible. |
the undivided Glossary of genetics article now exceeds the post-expand include size limit, so this page is being created to house approximately half of the original page's content, specifically those terms starting with numbers (0–9) through Z; this is a work-in-progress Tag: Disambiguation links added |
||
| Line 1: | Line 1: | ||
{{short description|List of definitions of terms and concepts commonly used in the study of genetics}} |
{{short description|List of definitions of terms and concepts commonly used in the study of genetics}} |
||
This '''glossary of genetics''' is a list of definitions of terms and concepts commonly used in the study of [[genetics]] and related disciplines in [[biology]], including [[molecular biology]], [[cell biology]], and [[evolutionary biology]].<ref>{{Cite web|url=https://www.genome.gov|title=Talking Glossary of Genetic Terms|date=8 October 2017|publisher=genome.gov|access-date=8 October 2017}}</ref> It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see [[Glossary of evolutionary biology]]. |
This '''glossary of genetics''' is a list of definitions of terms and concepts commonly used in the study of [[genetics]] and related disciplines in [[biology]], including [[molecular biology]], [[cell biology]], and [[evolutionary biology]].<ref>{{Cite web|url=https://www.genome.gov|title=Talking Glossary of Genetic Terms|date=8 October 2017|publisher=genome.gov|access-date=8 October 2017}}</ref> It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see [[Glossary of evolutionary biology]]. |
||
This glossary is split across two articles: |
|||
*'''Glossary of genetics (0–L)''' (this page) lists terms beginning with numbers and those beginning with the letters A through L. |
|||
*[[Glossary of genetics (M–Z)]] lists terms beginning with the letters M through Z. |
|||
__NOTOC__ |
__NOTOC__ |
||
| Line 1,349: | Line 1,353: | ||
{{term|lyonization}} |
{{term|lyonization}} |
||
{{defn|See ''{{gli|X-inactivation}}''.}} |
{{defn|See ''{{gli|X-inactivation}}''.}} |
||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==M== |
|||
{{glossary}} |
|||
{{term|[[major groove]]}} |
|||
{{defn|}} |
|||
<span id="map unit"></span>{{term|map unit (m.u.)}} |
|||
{{defn|See ''{{gli|centimorgan}}''.}} |
|||
{{term|map-based cloning}} |
|||
{{defn|See ''{{gli|positional cloning}}''.}} |
|||
{{term|matroclinous}} |
|||
{{defn|}} |
|||
{{term|[[medical genetics]]}} |
|||
{{defn|The branch of medicine and medical science that involves the study, diagnosis, and management of {{gli|genetic disorder|hereditary disorders}}, and more broadly the application of knowledge about human {{gli|genetics}} to medical care.}} |
|||
{{term|[[meiosis]]}} |
|||
{{defn|A specialized type of [[cell division]] that occurs exclusively in [[sexual reproduction|sexually reproducing]] [[eukaryote]]s, during which {{gli|DNA replication}} is followed by two consecutive rounds of division to ultimately produce four genetically unique {{gli|haploid}} daughter cells, each with half the number of {{gli|chromosomes}} as the original {{gli|diploid}} parent cell. Meiosis only occurs in cells of the sex organs, and serves the purpose of generating haploid {{gli|gametes}} such as [[sperm]], [[egg cell|eggs]], or [[spore]]s, which are later fused during [[fertilization]]. The two meiotic divisions, known as ''Meiosis I'' and ''Meiosis II'', may also include various {{gli|genetic recombination}} events between {{gli|homologous chromosomes}}.}} |
|||
{{term|meiotic spindle}} |
|||
{{defn|See ''{{gli|spindle apparatus}}''.}} |
|||
{{term|melting}} |
|||
{{defn|The {{gli|denaturation}} of a {{gli|double-stranded}} {{gli|nucleic acid}} into two {{gli|single strands}}, especially in the context of the {{gli|polymerase chain reaction}}.}} |
|||
{{term|[[Mendelian inheritance]]}} |
|||
{{defn|A theory of biological {{gli|heredity|inheritance}} based on a set of principles originally proposed by [[Gregor Mendel]] in 1865 and 1866. Mendel derived three generalized laws about the genetic basis of inheritance which, together with several theories developed by later scientists, are considered the foundation of {{gli|classical genetics}}. Contrast ''{{gli|non-Mendelian inheritance}}''.}} |
|||
<span id="mRNA"></span>{{term|[[messenger RNA]] (mRNA)}}{{anchor|messenger RNA|mRNAs}} |
|||
{{defn|Any of a class of {{gli|ssRNA|single-stranded}} {{gli|RNA}} molecules which function as molecular messengers, carrying sequence information encoded in the {{gli|DNA}} genome to the {{gli|ribosomes}} where protein synthesis occurs. The primary products of {{gli|transcription}}, mRNAs are synthesized by {{gli|RNA polymerase}}, which builds a chain of {{gli|ribonucleotides}} that complement the {{gli|deoxyribonucleotides}} of a {{gli|template strand|DNA template}}; in this way, the DNA sequence of a protein-coding {{gli|gene}} is effectively preserved in the {{gli|pre-mRNA|raw transcript}}, which is subsequently processed into a mature mRNA by a series of {{gli|post-transcriptional modifications}}.}} |
|||
[[File:MRNA_structure.svg|300px|thumb|right|The structure of a typical mature protein-coding '''{{gli|messenger RNA}}''' or '''mRNA''', drawn approximately to scale. The coding sequence (green) is bounded by {{gli|untranslated regions}} at both the {{gli|5' UTR|5'-end}} (yellow) and the {{gli|3' UTR|3'-end}} (pink). Prior to export from the nucleus, a {{gli|5' cap}} (red) and a {{gli|poly(A) tail|3' poly(A) tail}} (black) are added to help stabilize the mRNA and prevent its degradation by ribonucleases.]] |
|||
{{term|[[metabolome]]}} |
|||
{{defn|}} |
|||
{{term|[[centromere#metacentric|metacentric]]}} |
|||
{{defn|(of a linear {{gli|chromosome}} or chromosome fragment) Having a {{gli|centromere}} positioned in the middle of the chromosome, resulting in {{gli|chromatid}} arms of approximately equal length.<ref name="CoG"/>}} |
|||
{{term|[[metagenomics]]}} |
|||
{{ghat|Also '''environmental genomics''', '''ecogenomics''', and '''community genomics'''.}} |
|||
{{defn|The study of genetic material recovered directly from environmental samples, as opposed to organisms cultivated in [[microbiological culture|laboratory cultures]].}} |
|||
{{term|[[metaphase]]}} |
|||
{{defn|The stage of {{gli|mitosis}} and {{gli|meiosis}} that occurs after {{gli|prometaphase}} and before {{gli|anaphase}}, during which the {{gli|centromeres}} of the replicated chromosomes align along the equator of the cell, with each {{gli|kinetochore}} attached to the {{gli|mitotic spindle}}.}} |
|||
<span id="mAGE"></span>{{term|[[MicroArray and Gene Expression]] (MAGE)}} |
|||
{{defn|A group that "aims to provide a standard for the representation of {{gli|DNA microarray}} {{gli|gene expression}} data that would facilitate the exchange of microarray information between different data systems".<ref name="pmid17087822">{{cite journal | display-authors = 4| author = Rayner TF| author2 = Rocca-Serra P| author3 = Spellman PT| author4 = Causton HC| author5 = Farne A| author6 = Holloway E| author7 = Irizarry RA| author8 = Liu J| author9 = Maier DS| author10 = Miller M| author11 = Petersen K| author12 = Quackenbush J| author13 = Sherlock G| author14 = Stoeckert CJ| author15 = White J| author16 = Whetzel PL| author17 = Wymore F| author18 = Parkinson H| author19 = Sarkans U| author20 = Ball CA| author21 = Brazma A | title = A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB | journal = BMC Bioinformatics | volume = 7| pages = 489 | date = 2006 | pmid = 17087822 | pmc = 1687205 | doi = 10.1186/1471-2105-7-489 }}</ref>}} |
|||
{{term|[[microchromosome]]}} |
|||
{{defn|A type of very small {{gli|chromosome}}, generally less than 20,000 {{gli|base pairs}} in size, present in the {{gli|karyotypes}} of some organisms.}} |
|||
{{term|[[microdeletion]]}} |
|||
{{defn|A {{gli|chromosomal}} {{gli|deletion}} that is too short to cause any apparent change in morphology under a light microscope, though it may still be detectable with other methods such as {{gli|sequencing}}.}} |
|||
<span id="miRNA"></span>{{term|[[microRNA]] (miRNA)}} |
|||
{{defn|A type of small, {{gli|single-stranded RNA|single-stranded}}, {{gli|ncRNA|non-coding RNA}} molecule that functions in {{gli|post-transcriptional}} {{gli|gene regulation|regulation}} of {{gli|gene expression}}, particularly RNA {{gli|gene silencing|silencing}}, by base-pairing with complementary sequences in {{gli|mRNA|mRNA transcripts}}, which typically results in the cleavage or destabilization of the transcript or inhibits its {{gli|translation}} by ribosomes.}} |
|||
{{term|[[microsatellite]]}}{{anchor|microsatellites}} |
|||
{{ghat|Also '''short tandem repeat (STR)''' or '''simple sequence repeat (SSR)'''.}} |
|||
{{defn|A type of {{gli|satellite DNA}} consisting of a relatively short {{gli|nucleotide sequence|sequence}} of {{gli|tandem repeat|tandem repeats}}, in which certain {{gli|sequence motif|motifs}} (ranging in length from one to six or more {{gli|nitrogenous base|bases}}) are repeated, typically 5–50 times. Microsatellites are widespread throughout most organisms' {{gli|genome|genomes}} and tend to have higher mutation rates than other regions. They are classified as {{gli|variable number tandem repeat}} (VNTR) DNA, along with longer {{gli|minisatellite|minisatellites}}.}} |
|||
{{term|[[midparent value]]}} |
|||
{{defn|The mean of the two parental values for a {{gli|quantitative trait}} in an individual {{gli|offspring}} or in a specific {{gli|crossbreeding|cross}}.<ref name="DoG7"/>}} |
|||
<span id="mIAME"></span>{{term|[[MIAME|Minimum information about a microarray experiment]] (MIAME)}} |
|||
{{defn|A commercial standard developed by {{gli|FGED}} and based on {{gli|MAGE}} in order to facilitate the storage and sharing of {{gli|gene expression}} data.<ref name="pmid18629115">{{cite journal | author = Oliver S | title = On the MIAME Standards and Central Repositories of Microarray Data | journal = Comparative and Functional Genomics | volume = 4 | issue = 1 | pages = 1 | date = 2003 | pmid = 18629115 | pmc = 2447402 | doi = 10.1002/cfg.238 }}</ref><ref name="pmid19484163">{{cite journal | author = Brazma A | title = Minimum Information About a Microarray Experiment (MIAME)--successes, failures, challenges | journal = ScientificWorldJournal | volume = 9 | pages = 420–3 | date = 2009 | pmid = 19484163 | doi = 10.1100/tsw.2009.57 | pmc = 5823224 }}</ref>}} |
|||
<span id="mINSEQE"></span>{{term|[[MINSEQE|Minimal information about a high-throughput sequencing experiment]] (MINSEQE)}} |
|||
{{defn|A commercial standard developed by {{gli|FGED}} for the storage and sharing of {{gli|high-throughput}} {{gli|DNA sequencing|sequencing}} data.<ref name="minseqe">{{cite web | author = Functional Genomics Data Society | title = Minimum Information about a high-throughput SEQuencing Experiment |date=June 2012 | url = http://fged.org/projects/minseqe/ }}</ref>}} |
|||
{{term|[[minisatellite]]}}{{anchor|minisatellites}} |
|||
{{defn|}} |
|||
<span id="minor allele frequency"></span>{{term|[[minor allele frequency]] (MAF)}} |
|||
{{defn|}} |
|||
{{term|[[minor groove]]}} |
|||
{{defn|}} |
|||
{{term|minus-strand}} |
|||
{{defn|See ''{{gli|template strand}}''.}} |
|||
{{term|miRNA}} |
|||
{{defn|See ''{{gli|miRNA|microRNA}}''.}} |
|||
<span id="mismatch repair"></span>{{term|[[mismatch repair]] (MMR)}} |
|||
{{defn|}} |
|||
{{term|mismatch}}{{anchor|mismatches}} |
|||
{{ghat|Also '''mispairing'''.}} |
|||
{{defn|An incorrect {{gli|pairing}} of {{gli|nucleobases}} on {{gli|complementary}} {{gli|strands}} of {{gli|DNA}} or {{gli|RNA}}, i.e. the presence in one strand of a duplex molecule of a base that is not complementary (by Watson–Crick pairing rules) to the base occupying the corresponding position in the other strand, which prevents normal [[hydrogen bonding]] between the bases; e.g. a {{gli|guanine}} matched with a {{gli|thymine}}.<ref name="DoG7"/>}} |
|||
{{term|[[missense mutation]]}} |
|||
{{defn|A type of {{gli|point mutation}} which results in a {{gli|codon}} that codes for a different {{gli|amino acid}} than in the unmutated sequence. Compare ''{{gli|nonsense mutation}}''.}} |
|||
{{term|mistranslation}} |
|||
{{defn|The insertion of an incorrect {{gli|amino acid}} in a growing {{gli|peptide}} chain during {{gli|translation}}, i.e. the inclusion of any amino acid that is not the one specified by a particular {{gli|codon}} in an {{gli|mRNA}} transcript. Mistranslation may originate from a {{gli|mischarged}} {{gli|transfer RNA}} or from a malfunctioning {{gli|ribosome}}.<ref name="DoG7"/>}} |
|||
<span id="mtDNA"></span>{{term|[[mitochondrial DNA]] (mtDNA)}} |
|||
{{defn|}} |
|||
{{term|[[mitosis]]}}{{anchor|mitotic}} |
|||
{{defn|In [[eukaryote|eukaryotic]] [[cell (biology)|cells]], the part of the [[cell cycle]] during which the division of the {{gli|nucleus}} takes place and {{gli|replication|replicated}} {{gli|chromosomes}} are separated into two distinct nuclei. Mitosis is generally preceded by the S stage of {{gli|interphase}}, when the cell's {{gli|DNA}} is {{gli|DNA replication|replicated}}, and either occurs simultaneously with or is followed by [[cytokinesis]], when the [[cytoplasm]] and [[cell membrane]] are divided into two new daughter cells.}} |
|||
{{term|[[mitotic recombination]]}} |
|||
{{defn|}} |
|||
{{term|mitotic segregation}} |
|||
{{defn|}} |
|||
{{term|mitotic spindle}} |
|||
{{defn|See ''{{gli|spindle apparatus}}''.}} |
|||
{{term|[[mixoploidy]]}} |
|||
{{defn|The presence of more than one different {{gli|ploidy}} level, i.e. more than one number of sets of {{gli|chromosomes}}, in different cells of the same cellular population.<ref name="DoG7"/>}} |
|||
<span id="mobile genetic element"></span>{{term|[[mobile genetic element]] (MGE)}} |
|||
{{defn|Any genetic material that can move between different parts of a {{gli|genome}} or be transferred from one species or {{gli|replicon}} to another within a single {{gli|generation}}. The many types of MGEs include {{gli|transposable elements}}, bacterial {{gli|plasmids}}, [[bacteriophage]] elements which integrate into host genomes by viral {{gli|transduction}}, and {{gli|self-splicing introns}}.}} |
|||
{{term|[[mobilome]]}} |
|||
{{defn|The entire set of {{gli|mobile genetic elements}} within a particular {{gli|genome}}, cell, species, or other taxon, including all {{gli|transposons}}, {{gli|plasmids}}, {{gli|prophages}}, and other self-splicing nucleic acid molecules.}} |
|||
{{term|[[molecular cloning]]}} |
|||
{{defn|Any of various {{gli|molecular biology}} methods designed to {{gli|replicate}} a particular molecule, usually a DNA sequence, many times inside the cells of a natural host. Commonly, a {{gli|recombinant DNA}} fragment containing a {{gli|gene of interest}} is {{gli|ligation|ligated}} into a {{gli|plasmid}} {{gli|vector}}, which is then {{gli|transformed}} into {{gli|competent}} bacterial cells. The bacteria, carrying the recombinant plasmid, are then allowed to proliferate naturally on a culture medium, where the plasmids are replicated along with the rest of the bacterial genome; any functioning gene of interest will be {{gli|expressed}} by the bacterial cells, and thereby its {{gli|gene products}} will also be cloned. Molecular cloning is a fundamental tool of {{gli|genetic engineering}}, designed to study gene expression, to amplify a specific gene product, and to generate a {{gli|selectable marker|selectable phenotype}}.}} |
|||
[[File:Gene_cloning.svg|350px|thumb|right|An outline of how '''{{gli|molecular cloning}}''' works]] |
|||
{{term|[[molecular genetics]]}} |
|||
{{defn|A branch of {{gli|genetics}} that employs methods and techniques of {{gli|molecular biology}} to study the structure and function of {{gli|genes}} and {{gli|gene product|gene products}} at the [[molecule|molecular]] level. Contrast ''{{gli|classical genetics}}''.}} |
|||
{{term|monad}} |
|||
{{defn|A {{gli|haploid}} set of {{gli|chromosomes}} as it exists inside the {{gli|nucleus}} of an immature {{gli|gamete|gametic cell}} such as an [[ootid]] or [[spermatid]], i.e. a cell which has completed {{gli|meiosis}} but is not yet a mature gamete.<ref name="DoG7"/>}} |
|||
{{term|[[monocentric]]}} |
|||
{{defn|(of a linear {{gli|chromosome}} or chromosome fragment) Having only one {{gli|centromere}}. Contrast ''{{gli|dicentric}}'' and ''{{gli|holocentric}}''.}} |
|||
{{term|[[monoploid]]}} |
|||
{{defn|}} |
|||
{{term|[[monosomy]]}} |
|||
{{defn|The abnormal and frequently pathological presence of only one {{gli|chromosome}} of a normal diploid pair. It is a type of {{gli|aneuploidy}}.}} |
|||
{{term|[[Morpholino]]}} |
|||
{{ghat|Also '''phosphorodiamidate Morpholino oligomer'''.}} |
|||
{{defn|A synthetic polymeric molecule connecting short sequences of {{gli|nucleobases}} into artificial {{gli|antisense}} {{gli|oligomer|oligomers}}, which are used in {{gli|genetic engineering}} to {{gli|knockdown}} {{gli|gene expression}} by {{gli|base pair|pairing}} with {{gli|complementarity|complementary}} sequences in naturally occurring RNA or DNA molecules, especially {{gli|mRNA|mRNA transcripts}}, thereby blocking access by other biomolecules such as {{gli|ribosome|ribosomes}} and proteins. Morpholino oligomers are not themselves {{gli|translated}}, and neither they nor their hybrid duplexes with RNA are attacked by {{gli|nucleases}}; also, unlike the negatively charged {{gli|phosphate backbone|phosphates}} of normal nucleic acids, the synthetic backbones of Morpholinos are electrically neutral, making them less likely to interact non-selectively with a host cell's charged proteins. These properties make them useful and reliable tools for artificially generating {{gli|mutant}} phenotypes in living cells.<ref name="DoG7"/>}} |
|||
{{term|[[mosaic (genetics)|mosaicism]]}} |
|||
{{defn|The presence of two or more populations of cells with different {{gli|genotypes}} in an individual organism which has developed from a single fertilized [[ovum|egg]]. A mosaic organism can result from many kinds of genetic phenomena, including {{gli|nondisjunction}} of chromosomes, {{gli|endoreduplication}}, or mutations in individual [[stem cell]] lineages during the early development of the embryo. Mosaicism is similar to but distinct from {{gli|chimerism}}.}} |
|||
{{term|[[sequence motif|motif]]}}{{anchor|motifs}} |
|||
{{defn|Any distinctive or recurring {{gli|sequence}} of {{gli|nucleotides}} in a {{gli|nucleic acid}} or of {{gli|amino acids}} in a {{gli|protein}} that is or is conjectured to be biologically significant, especially one that is reliably {{gli|recognition site|recognized}} by other biomolecules or which has a {{gli|secondary structure|three-dimensional structure}} that permits unique or characteristic chemical interactions such as {{gli|DNA binding}}.<ref name="DoG7"/> In nucleic acids, motifs are often short (three to ten nucleotides in length), highly {{gli|conserved sequence|conserved sequences}} which act as recognition sites for {{gli|DNA-binding protein|DNA-binding enzymes}} or RNAs involved in the regulation of {{gli|gene expression}}.}} |
|||
{{term|mRNA}} |
|||
{{defn|See ''{{gli|mRNA|messenger RNA}}''.}} |
|||
{{term|mtDNA}} |
|||
{{defn|See ''{{gli|mtDNA|mitochondrial DNA}}''.}} |
|||
{{term|[[multiomics]]}} |
|||
{{defn|}} |
|||
<span id="multiple cloning site"></span>{{term|[[multiple cloning site]] (MCS)}}{{anchor|polylinker}} |
|||
{{ghat|Also '''polylinker'''.}} |
|||
{{defn|}} |
|||
{{term|[[mutagen]]}}{{anchor|mutagens}} |
|||
{{defn|Any physical or chemical agent that {{gli|mutagenesis|changes the genetic material}} (usually {{gli|DNA}}) of an organism and thereby increases the frequency of {{gli|mutations}} above natural background levels.}} |
|||
{{term|[[mutagenesis]]}} |
|||
{{defn|no=1|The process by which the genetic information of an organism is changed, resulting in a {{gli|mutation}}. Mutagenesis may occur spontaneously or as a result of exposure to a {{gli|mutagen}}.}} |
|||
{{defn|no=2|In {{gli|molecular biology}}, any laboratory technique by which one or more genetic mutations are deliberately {{gli|genetic engineering|engineered}} in order to produce a {{gli|mutant}} gene, regulatory element, gene product, or {{gli|genetically modified organism}} so that the functions of a genetic locus, process, or product can be studied in detail.}} |
|||
{{term|[[mutant]]}}{{anchor|mutants}} |
|||
{{defn|An organism, gene product, or phenotypic {{gli|trait}} resulting from a {{gli|mutation}}, of a type that would not be observed naturally in {{gli|wild-type}} specimens.}} |
|||
{{term|[[mutation]]}}{{anchor|mutations|mutate|mutates|mutated}} |
|||
{{defn|Any permanent change in the {{gli|nucleotide sequence}} of a strand of {{gli|DNA}} or {{gli|RNA}}, or in the {{gli|amino acid}} sequence of a {{gli|peptide}}. Mutations play a role in both normal and abnormal biological processes; their natural occurrence is integral to the process of {{gli|evolution}}. They can result from errors in {{gli|DNA replication|replication}}, chemical damage, exposure to high-energy radiation, or manipulations by {{gli|mobile genetic element|mobile genetic elements}}. {{gli|DNA repair|Repair}} mechanisms have evolved in many organisms to correct them. By understanding the effect that a mutation has on {{gli|phenotype}}, it is possible to establish the function of the {{gli|gene}} or sequence in which it occurs.}} |
|||
{{term|mutation distance}} |
|||
{{defn|The smallest number of {{gli|mutations}} required to derive one particular gene, sequence, or phenotype from another;<ref name="DoG7"/> the minimum number of nucleobase insertions, deletions, or substitutions necessary to change one sequence into another.}} |
|||
{{term|[[mutation event]]}} |
|||
{{defn|The actual origin of a particular {{gli|mutation}} in time and space, the instance of its original introduction into a {{gli|genome}}, as opposed to that of its phenotypic manifestation, which may only occur {{gli|generations}} after the fact.}} |
|||
{{term|[[mutation rate]]}} |
|||
{{defn|The frequency of new {{gli|mutations}} at a particular {{gli|locus}} or in a particular {{gli|gene}}, {{gli|nucleotide sequence|sequence}}, {{gli|genome}}, or organism over a specified period of time, e.g. during a single {{gli|generation}}. Mutation rates may be calculated for a specific class of mutation or for all types collectively; they vary widely by organism and with an organism's environment.}} |
|||
{{term|mutational hot spot}} |
|||
{{defn|}} |
|||
{{term|[[mutational load]]}} |
|||
{{defn|See ''{{gli|genetic load}}''.}} |
|||
{{term|mutator gene}} |
|||
{{defn|Any mutant {{gli|gene}} or sequence that increases the spontaneous {{gli|mutation rate}} of one or more other genes or sequences. Mutators are often {{gli|transposable element|transposable elements}}, or may be mutant {{gli|housekeeping gene|housekeeping genes}} such as those that encode {{gli|helicase|helicases}} or proteins involved in {{gli|DNA proofreading|proofreading}}.<ref name="DoG7"/>}} |
|||
{{term|[[mutein]]}} |
|||
{{defn|A {{gli|mutant}} {{gli|protein}}, i.e. a protein whose {{gli|amino acid}} sequence differs from that of the normal because of a {{gli|mutation}}.}} |
|||
{{term|muton}} |
|||
{{defn|The smallest unit of a DNA molecule in which a physical or chemical change can result in a {{gli|mutation}} (conventionally a single {{gli|nucleotide}}).<ref name="DoG7"/>}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==N== |
|||
{{glossary}} |
|||
{{term|''n'' orientation}} |
|||
{{defn|One of two possible orientations by which a linear DNA fragment can be inserted into a {{gli|vector}}, specifically the one in which the {{gli|gene map|gene maps}} of both fragment and vector have the same orientation.<ref name="DoG7"/> Contrast ''{{gli|u orientation}}''.}} |
|||
{{term|[[N-terminus]]}}{{anchor|N-terminal}} |
|||
{{ghat|Also '''amine terminus''' and '''amino terminus'''.}} |
|||
{{defn|The end of a linear chain of {{gli|amino acids}} (i.e. a {{gli|peptide}}) that is terminated by the free [[amine group]] ({{chem|-NH|2}}) of the first amino acid added to the chain during {{gli|translation}}. This amino acid is said to be ''N-terminal''. By convention, sequences, domains, active sites, or any other structure positioned nearer to the N-terminus of the {{gli|polypeptide}} or its folded protein form relative to others are described as {{gli|upstream}}. Contrast ''{{gli|C-terminus}}''.}} |
|||
{{term|NAD}} |
|||
{{defn|See ''{{gli|nicotinamide-adenine dinucleotide}}''.}} |
|||
{{term|NADP}} |
|||
{{defn|See ''{{gli|nicotinamide-adenine dinucleotide phosphate}}''.}} |
|||
{{term|nascent}} |
|||
{{defn|In the process of being synthesized; incomplete; not yet fully processed or mature. The term is commonly used to describe {{gli|strand|strands}} of {{gli|DNA}} or {{gli|RNA}} which are actively undergoing synthesis during {{gli|DNA replication|replication}} or {{gli|transcription}}, respectively, or sometimes a complete, fully transcribed RNA molecule before any {{gli|post-transcriptional modification|alterations}} have been made (e.g. {{gli|polyadenylation}} or {{gli|RNA editing}}), or a {{gli|peptide}} chain actively undergoing {{gli|translation}} by a {{gli|ribosome}}.<ref name="DoG7"/>}} |
|||
{{term|ncDNA}} |
|||
{{defn|See ''{{gli|ncDNA|non-coding DNA}}''.}} |
|||
{{term|ncRNA}} |
|||
{{defn|See ''{{gli|ncRNA|non-coding RNA}}''.}} |
|||
{{term|negative control}} |
|||
{{ghat|Also '''negative regulation'''.}} |
|||
{{defn|A type of {{gli|gene regulation}} in which a {{gli|repressor}} binds to an {{gli|operator}} upstream from the coding region and thereby prevents {{gli|transcription}} by {{gli|RNA polymerase}}. An {{gli|inducer}} is necessary to switch on transcription in {{gli|positive control}}.<ref name="Rieger"/>}} |
|||
{{term|negative (-) sense strand}} |
|||
{{defn|See ''{{gli|template strand}}''.}} |
|||
{{term|[[negative supercoiling]]}} |
|||
{{defn|}} |
|||
{{term|[[neutral mutation]]}} |
|||
{{defn|no=1|Any {{gli|mutation}} of a {{gli|nucleic acid sequence}} that is neither beneficial nor detrimental to the ability of an organism to survive and reproduce.}} |
|||
{{defn|no=2|Any mutation for which [[natural selection]] does not affect the spread of the mutation within a population.}} |
|||
{{term|[[nick (DNA)|nick]]}}{{anchor|nicks|nicking|nicked}} |
|||
{{defn|}} |
|||
{{term|[[nick translation]]}} |
|||
{{defn|}} |
|||
{{term|[[nicking enzyme]]}} |
|||
{{defn|}} |
|||
<span id="NAD"></span>{{term|[[nicotinamide adenine dinucleotide]] (NAD)}} |
|||
{{defn|}} |
|||
<span id="NADP"></span>{{term|[[nicotinamide adenine dinucleotide phosphate]] (NADP, NADP+)}} |
|||
{{defn|}} |
|||
{{term|[[nitrogenous base]]}}{{anchor|nitrogenous bases}} |
|||
{{ghat|Sometimes used interchangeably with '''{{gli|nucleobase}}''' or simply '''base'''.}} |
|||
{{defn|Any organic compound containing a [[nitrogen]] atom that has the chemical properties of a [[base (chemistry)|base]]. A set of {{gli|nucleobase|five particular nitrogenous bases}} – {{gli|adenine}} ({{font|A|font=courier|size=big}}), {{gli|guanine}} ({{font|G|font=courier|size=big}}), {{gli|cytosine}} ({{font|C|font=courier|size=big}}), {{gli|thymine}} ({{font|T|font=courier|size=big}}), and {{gli|uracil}} ({{font|U|font=courier|size=big}}) – are especially relevant to biology because they are components of {{gli|nucleotides}}, which in turn are the primary [[monomer]]s that make up {{gli|nucleic acids}}.}} |
|||
<span id="ncDNA"></span>{{term|[[non-coding DNA]] (ncDNA)}} |
|||
{{defn|Any segment of {{gli|DNA}} that does not {{gli|genetic code|encode}} a sequence that may ultimately be {{gli|transcribed}} and {{gli|translated}} into a {{gli|protein}}. In most organisms, only a small fraction of the genome consists of protein-coding DNA, though the proportion varies greatly between species. Some non-coding DNA may still be transcribed into functional {{gli|ncRNA|non-coding RNA}} (as with {{gli|tRNA|transfer RNAs}}) or may serve important developmental or {{gli|gene regulation|regulatory}} purposes; other regions (as with so-called "{{gli|junk DNA}}") appear to have no known biological function.}} |
|||
<span id="ncRNA"></span>{{term|[[non-coding RNA]] (ncRNA)}}{{anchor|non-coding RNA|non-coding RNAs}} |
|||
{{defn|Any molecule of {{gli|RNA}} that is not ultimately {{gli|translated}} into a {{gli|protein}}. The {{gli|DNA}} sequence from which a functional non-coding RNA is {{gli|transcribed}} is often referred to as an "RNA gene". Numerous types of non-coding RNAs essential to normal genome function are produced constitutively, including {{gli|tRNA|transfer RNA}} (tRNA), {{gli|rRNA|ribosomal RNA}} (rRNA), {{gli|miRNA|microRNA}} (miRNA), and {{gli|siRNA|small interfering RNA}} (siRNA); other non-coding RNAs (sometimes described as "junk RNA") have no known function and are likely the product of spurious transcription.}} |
|||
<span id="non-homologous end joining"></span>{{term|[[non-homologous end joining]] (NHEJ)}} |
|||
{{defn|}} |
|||
{{term|[[non-Mendelian inheritance]]}} |
|||
{{defn|Any pattern of {{gli|inheritance}} in which traits do not segregate in accordance with {{gli|Mendelian inheritance|Mendel's laws}}, which describe the readily observable inheritance of discretely variable phenotypic traits influenced by single {{gli|genes}} on nuclear {{gli|chromosomes}}. Though they correctly explain many basic observations of inheritance, Mendel's laws are useful only in the simplest and most general cases; there exist numerous genetic processes and phenomena, both normal and abnormal, which violate them, such as {{gli|incomplete dominance}}, {{gli|codominance}}, {{gli|linkage|genetic linkage}}, {{gli|epistatic}} interactions and {{gli|polygene|polygenic traits}}, {{gli|non-random segregation of chromosomes}}, {{gli|extranuclear inheritance}}, {{gli|gene conversion}}, and many {{gli|epigenetic}} phenomena.}} |
|||
{{term|[[non-random segregation of chromosomes]]}} |
|||
{{defn|}} |
|||
{{term|non-transcribed spacer (NTS)}} |
|||
{{defn|See ''{{gli|spacer}}''.}} |
|||
{{term|noncoding strand}} |
|||
{{defn|See ''{{gli|template strand}}''.}} |
|||
{{term|[[nondisjunction]]}} |
|||
{{defn|The failure of {{gli|homologous chromosomes}} or {{gli|sister chromatids}} to {{gli|segregate}} properly during [[cell division]]. Nondisjunction results in daughter cells that are {{gli|aneuploid}}, containing abnormal numbers of one or more specific {{gli|chromosomes}}. It may be caused by any of a variety of factors.}} |
|||
{{term|[[nonhomologous recombination]]}} |
|||
{{defn|}} |
|||
{{term|nonreciprocal recombination}} |
|||
{{defn|See ''{{gli|unequal crossing over}}''.}} |
|||
{{term|nonrepetitive sequence}} |
|||
{{defn|Broadly, any {{gli|nucleotide sequence}} or region of a {{gli|genome}} that does not contain {{gli|repeated sequences}}, or in which repeats do not comprise a majority; or any segment of DNA exhibiting the {{gli|reassociation kinetics}} expected of a unique sequence.<ref name="DoG7"/>}} |
|||
{{term|[[nonsense mutation]]}} |
|||
{{ghat|Also '''point-nonsense mutation'''.}} |
|||
{{defn|A type of {{gli|point mutation}} which results in a premature {{gli|stop codon}} in the {{gli|transcribed}} {{gli|mRNA}} sequence, thereby causing the premature termination of {{gli|translation}}, which results in a truncated, incomplete, and often non-functional {{gli|protein}}.}} |
|||
{{term|[[nonsense suppressor]]}} |
|||
{{defn|}} |
|||
{{term|[[nonsynonymous substitution|nonsynonymous mutation]]}} |
|||
{{ghat|Also '''nonsynonymous substitution''' or '''replacement mutation'''.}} |
|||
{{defn|A type of {{gli|mutation}} in which the {{gli|substitution}} of one {{gli|nucleotide}} base for another results, after {{gli|transcription}} and {{gli|translation}}, in an amino acid sequence that is different from that produced by the original unmutated gene. Because nonsynonymous mutations always result in a biological change in the organism, they are often subject to strong {{gli|natural selection|selection pressure}}. Contrast ''{{gli|synonymous mutation}}''.}} |
|||
{{term|[[Northern blotting]]}} |
|||
{{defn|}} |
|||
{{term|nRNA}} |
|||
{{defn|See ''{{gli|nuclear RNA}}''.}} |
|||
{{term|[[nuclear membrane]]}} |
|||
{{ghat|Also '''nuclear envelope'''.}} |
|||
{{defn|A sub-cellular barrier consisting of two [[lipid bilayer]] [[polar membrane|membranes]] that surrounds the {{gli|nucleus}} in [[eukaryote|eukaryotic cells]].}} |
|||
<span id="nuclear RNA"></span>{{term|nuclear RNA (nRNA)}} |
|||
{{defn|Any {{gli|RNA}} molecule located within a cell's {{gli|nucleus}}, whether associated with {{gli|chromosomes}} or existing freely in the nucleoplasm, including {{gli|snRNA|small nuclear RNA}} (snRNA), {{gli|eRNA|enhancer RNA}} (eRNA), and all newly transcribed {{gli|hnRNA|immature RNAs}}, {{gli|mRNA|coding}} or {{gli|ncRNA|non-coding}}, prior to their export to the cytosol (hnRNA).}} |
|||
{{term|[[nuclease]]}}{{anchor|nucleases}} |
|||
{{defn|Any of a class of enzymes capable of cleaving {{gli|phosphodiester bonds}} between adjacent nucleotides in a nucleic acid molecule (the opposite of a {{gli|ligase}}). Nucleases may {{gli|nick|nick one}} or {{gli|cut|cut both}} {{gli|strands}} of a duplex molecule; they may cleave randomly or at specific recognition sequences. They are ubiquitous and imperative for normal cellular function, and are also widely employed in {{gli|molecular biology}} techniques.}} |
|||
{{term|[[nucleic acid]]}}{{anchor|nucleic acids}} |
|||
{{defn|A long, [[polymer]]ic [[macromolecule]] made up of smaller [[monomer]]s called {{gli|nucleotides}} which are chemically linked to one another in a chain. Two specific types of nucleic acid, {{gli|DNA}} and {{gli|RNA}}, are used in biological systems to encode the genetic information governing the construction, development, and ordinary processes of all living organisms. The order, or {{gli|sequence}}, of the nucleotides in DNA and RNA molecules contains information that is translated into {{gli|proteins}}, which direct all of the chemical reactions necessary for life.}} |
|||
{{term|[[nucleic acid sequence]]}}{{anchor|nucleic acid sequences|nucleotide sequence|nucleotide sequences|sequence|sequences|DNA sequence|DNA sequences}} |
|||
{{defn|The precise order of consecutively linked {{gli|nucleotides}} in a {{gli|nucleic acid}} molecule such as {{gli|DNA}} or {{gli|RNA}}. Long sequences of nucleotides are the principal means by which biological systems store genetic information, and therefore the accurate {{gli|replication}}, {{gli|transcription}}, and {{gli|translation}} of such sequences is of the utmost importance, lest the information be lost or corrupted. Nucleic acid sequences may be equivalently referred to as sequences of {{gli|nitrogenous bases}}, {{gli|nucleobases}}, nucleotides, or {{gli|base pairs}}, and they correspond directly to sequences of {{gli|codons}} and {{gli|amino acids}}.}} |
|||
{{term|[[nucleobase]]}}{{anchor|base|bases|nucleobases}} |
|||
{{ghat|Sometimes used interchangeably with '''{{gli|nitrogenous base}}''' or simply '''base'''.}} |
|||
{{defn|One of the five primary or canonical {{gli|nitrogenous base|nitrogenous bases}} – {{gli|adenine}} ({{font|A|font=courier|size=big}}), {{gli|guanine}} ({{font|G|font=courier|size=big}}), {{gli|cytosine}} ({{font|C|font=courier|size=big}}), {{gli|thymine}} ({{font|T|font=courier|size=big}}), and {{gli|uracil}} ({{font|U|font=courier|size=big}}) – that form {{gli|nucleosides}} and {{gli|nucleotides}}, the latter of which are the fundamental building blocks of {{gli|nucleic acids}}. The ability of these nucleobases to form {{gli|base pairs}} via [[hydrogen bonding]], as well as their flat, compact three-dimensional profiles, allows them to "stack" one upon another and leads directly to the long-chain structures of {{gli|DNA}} and {{gli|RNA}}.}} |
|||
{{term|[[nucleoid]]}}{{anchor|nucleoids}} |
|||
{{ghat|Also '''prokaryon'''.}} |
|||
{{defn|An irregularly shaped region within a prokaryotic cell which contains most or all of the cell's genetic material, but is not enclosed by a {{gli|nuclear membrane}} as in eukaryotes.}} |
|||
{{term|[[nucleolus]]}}{{anchor|nucleoli}} |
|||
{{defn|An [[organelle]] within the {{gli|nucleus}} of [[eukaryote|eukaryotic]] cells which is composed of proteins, DNA, and RNA and serves as the site of {{gli|ribosome}} synthesis.}} |
|||
{{term|[[nucleoprotein]]}}{{anchor|nucleoproteins}} |
|||
{{defn|Any {{gli|protein}} that is chemically bonded to or conjugated with a {{gli|nucleic acid}} molecule. Examples include {{gli|ribosomes}}, {{gli|nucleosomes}}, and many enzymes.}} |
|||
{{term|[[nucleoside]]}}{{anchor|nucleosides}} |
|||
{{defn|An organic molecule composed of a {{gli|nitrogenous base}} bonded to a [[pentose|five-carbon sugar]] (either {{gli|ribose}} or {{gli|deoxyribose}}). A {{gli|nucleotide}} additionally includes one or more [[phosphate|phosphate groups]].}} |
|||
{{term|[[nucleosome]]}}{{anchor|nucleosomes}} |
|||
{{defn|}} |
|||
{{term|[[nucleotide]]}}{{anchor|nucleotides}} |
|||
{{ghat|Also '''nucleoside monophosphate (NMP)'''.}} |
|||
{{defn|An organic molecule that serves as the [[monomer]] or subunit of {{gli|nucleic acid}} polymers, including {{gli|RNA}} and {{gli|DNA}}. Each nucleotide is composed of three connected functional groups: a {{gli|nitrogenous base}}, a [[pentose|five-carbon sugar]] (either {{gli|ribose}} or {{gli|deoxyribose}}), and a single [[phosphate|phosphate group]]. Though technically distinct, the term "nucleotide" is often used interchangeably with nitrogenous base, {{gli|nucleobase}}, and {{gli|base pair}} when referring to the {{gli|sequences}} that make up nucleic acids. Compare ''{{gli|nucleoside}}''.}} |
|||
[[File:Nucleotides_1.svg|350px|thumb|right|The '''{{gli|nucleobases}}''' (blue) are the five specific {{gli|nitrogenous bases}} canonically used in DNA and RNA. A nucleobase bonded to a pentose sugar (either {{gli|ribose}} or {{gli|deoxyribose}}; yellow) is known as a '''{{gli|nucleoside}}''' (yellow + blue). A nucleoside bonded to a single phosphate group (red) is known as a nucleoside monophosphate (NMP) or a '''{{gli|nucleotide}}''' (red + yellow + blue). When not incorporated into a nucleic acid chain, free nucleosides can bind multiple phosphate groups: two phosphates yields a {{gli|nucleoside diphosphate}} (NDP), and three yields a {{gli|nucleoside triphosphate}} (NTP).]] |
|||
{{term|[[cell nucleus|nucleus]]}}{{anchor|nuclei|nuclear}} |
|||
{{ghat|Plural '''nuclei'''.}} |
|||
{{defn|A {{gli|nuclear membrane|membrane}}-enclosed [[organelle]] found in [[eukaryote|eukaryotic]] cells which contains most of the cell's genetic material (organized as {{gli|chromosomes}}) and directs the activities of the cell by regulating {{gli|gene expression}}.}} |
|||
{{term|[[null allele]]}} |
|||
{{defn|Any {{gli|allele}} made non-functional by way of a genetic {{gli|mutation}}. The mutation may result in the complete failure to produce a {{gli|gene product}} or a gene product that does not function properly; in either case, the allele may be considered non-functional.}} |
|||
{{term|[[zygosity#nullizygous|nullizygous]]}} |
|||
{{defn|}} |
|||
{{term|[[nullisomic]]}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==O== |
|||
{{glossary}} |
|||
{{term|[[stop codon#nomenclature|ochre]]}} |
|||
{{defn|One of three {{gli|stop codon|stop codons}} used in the {{gli|standard genetic code}}; in {{gli|RNA}}, it is specified by the nucleotide triplet {{font|UAA|font=courier|size=big}}. The other two stop codons are named {{gli|amber}} and {{gli|opal}}.}} |
|||
{{term|[[Okazaki fragments]]}} |
|||
{{defn|{{gli|oligonucleotide|Short sequences}} of {{gli|nucleotides}} which are synthesized discontinuously by {{gli|DNA polymerase}} and later linked together by {{gli|DNA ligase}} to create the {{gli|lagging strand}} during {{gli|DNA replication}}. Okazaki fragments are the consequence of the unidirectionality of DNA polymerase, which only works in the 5' to 3' direction.}} |
|||
{{term|[[oligogene]]}} |
|||
{{defn|}} |
|||
{{term|[[oligomer]]}} |
|||
{{defn|Any [[polymer]]ic molecule consisting of a relatively short series of connected [[monomer]]s or subunits; e.g. an {{gli|oligonucleotide}} is a short series of nucleotides.}} |
|||
{{term|[[oligonucleotide]]}}{{anchor|oligonucleotides|oligo|oligos}} |
|||
{{ghat|Also abbreviated '''oligo'''.}} |
|||
{{defn|A relatively short chain of {{gli|nucleic acid}} {{gli|residue|residues}}. In the laboratory, oligonucleotides are commonly used as {{gli|primers}} or {{gli|hybridization probes}} to detect the presence of larger {{gli|mRNA}} molecules or assembled into two-dimensional [[microarray]]s for {{gli|high-throughput}} {{gli|sequencing}} analysis.}} |
|||
{{term|omnipotent suppressor}} |
|||
{{defn|}} |
|||
{{term|[[oncogene]]}} |
|||
{{defn|A {{gli|gene}} that has the potential to cause [[cancer]]. In [[neoplasm|tumor]] cells, such genes are often {{gli|mutation|mutated}} and/or {{gli|expressed}} at abnormally high levels.}} |
|||
{{term|one gene–one polypeptide}} |
|||
{{ghat|Also '''one gene–one protein''' or '''one gene–one enzyme'''.}} |
|||
{{defn|The hypothesis that there exists a large class of {{gli|genes}} in which each particular gene directs the synthesis of one particular {{gli|polypeptide}} or {{gli|protein}}.<ref name="DoG7"/> Historically it was thought that all genes and proteins might follow this rule by definition, but it is now known that many or most proteins are composites of different polypeptides and therefore the product of multiple genes, and also that some genes do not encode polypeptides at all but instead produce {{gli|ncRNA|non-coding RNAs}}, which are never translated.}} |
|||
{{term|[[stop codon#nomenclature|opal]]}} |
|||
{{ghat|Also '''umber'''.}} |
|||
{{defn|One of three {{gli|stop codon|stop codons}} used in the {{gli|standard genetic code}}; in {{gli|RNA}}, it is specified by the nucleotide triplet {{font|UGA|font=courier|size=big}}. The other two stop codons are named {{gli|amber}} and {{gli|ochre}}.}} |
|||
{{term|[[open chromatin]]}} |
|||
{{defn|See ''{{gli|euchromatin}}''.}} |
|||
<span id="open reading frame"></span>{{term|[[open reading frame]] (ORF)}}{{anchor|open reading frames|ORF|ORFs}} |
|||
{{defn|The part of a {{gli|reading frame}} that has the ability to be {{gli|translated}} from DNA or RNA into protein; any continuous stretch of {{gli|codons}} that contains a {{gli|start codon}} and a {{gli|stop codon}}.}} |
|||
{{term|[[operator (genetics)|operator]]}}{{anchor|operators}} |
|||
{{defn|A regulatory sequence within an {{gli|operon}}, typically located between the {{gli|promoter}} sequence and the {{gli|structural genes}} of the operon, to which an uninhibited {{gli|repressor}} protein can bind, thereby physically obstructing {{gli|RNA polymerase}} from initiating the {{gli|transcription}} of adjacent cistrons.<ref name="Lewin"/>}} |
|||
{{term|[[operon]]}}{{anchor|operons}} |
|||
{{defn|A functional unit of {{gli|gene expression}} consisting of a cluster of adjacent {{gli|structural genes}} which are collectively under the control of a single {{gli|promoter}}, along with one or more adjacent {{gli|regulatory}} sequences such as {{gli|operators}} which affect {{gli|transcription}} of the structural genes. The set of genes is transcribed together, usually resulting in a single {{gli|polycistronic}} {{gli|mRNA|messenger RNA}} molecule, which may then be {{gli|translated}} together or undergo {{gli|RNA splicing|splicing}} to create multiple mRNAs which are translated independently; the result is that the genes contained in the operon are either expressed together or not at all. Regulatory proteins, including {{gli|repressors}}, {{gli|corepressors}}, and {{gli|activators}}, usually bind specifically to the regulatory sequences of a given operon; by some definitions, the genes that code for these regulatory proteins are also considered part of the operon.}} |
|||
{{term|operon network}} |
|||
{{defn|}} |
|||
<span id="origin of replication"></span>{{term|[[origin of replication]] (ORI)}}{{anchor|replication origin|origin}} |
|||
{{defn|}} |
|||
{{term|[[orphan gene]]}} |
|||
{{defn|A {{gli|gene}} for which there are no known functional homologs outside of a given species or {{gli|lineage}}, and whose evolutionary history is therefore obscure.}} |
|||
{{term|[[orphon]]}} |
|||
{{defn|}} |
|||
{{term|[[ortholog]]}}{{anchor|orthologs|orthologous}} |
|||
{{defn|One of a set of {{gli|genes}} (or more generally any DNA sequences showing {{gli|sequence homology|homology}}) which are present in different {{gli|genomes}} but are directly related to one another by vertical descent from a single gene or sequence in the [[last common ancestor]] of those genomes; such genes or sequences are said to be ''orthologous''. Orthologs are descended from the same ancestral sequence and can be inferred to be related to each other based on the similarity of their sequences; though they may have evolved independently within the separate genomes by mutation and natural selection, their {{gli|gene product|products}} may still retain similar structures, functions, or levels of {{gli|expression}} across species and populations. The identification of orthologs has proven important in inferring {{gli|phylogenetic}} relationships between different organisms. Contrast ''{{gli|paralog}}''.}} |
|||
{{term|[[outbreeding]]}} |
|||
{{ghat|Also '''outcrossing''' or '''{{gli|crossbreeding}}'''.}} |
|||
{{defn|[[Sexual reproduction]] between different breeds or individuals, which has the potential to increase {{gli|genetic diversity}} by introducing unrelated genetic material into a breeding population. The reproductive event and the resulting progeny may both be referred to as an ''outcross'', and the progeny is said to be ''outbred''. Contrast ''{{gli|inbreeding}}''.}} |
|||
{{term|[[outron]]}} |
|||
{{defn|A sequence near the {{gli|5'-end}} of a {{gli|primary transcript|primary mRNA transcript}} that is removed by a special form of {{gli|RNA splicing|splicing}} during {{gli|post-transcriptional modification|post-transcriptional processing}}. Outrons are located entirely outside of the transcript's {{gli|exon|coding sequences}}, unlike {{gli|introns}}.}} |
|||
{{term|overexpression}} |
|||
{{defn|An abnormally high level of {{gli|gene expression}} which results in an excessive number of copies of one or more {{gli|gene product|gene products}}. Overexpression produces a pronounced gene-related {{gli|phenotype}}.<ref name="Overexpression - Oxford">{{cite web|title=overexpression|url=https://en.oxforddictionaries.com/definition/overexpression|archive-url=https://web.archive.org/web/20180210181025/https://en.oxforddictionaries.com/definition/overexpression|url-status=dead|archive-date=February 10, 2018|website=Oxford Living Dictionary|publisher=Oxford University Press|access-date=18 May 2017|date=2017|quote=The production of abnormally large amounts of a substance which is coded for by a particular gene or group of genes; the appearance in the phenotype to an abnormally high degree of a character or effect attributed to a particular gene.}}</ref><ref name="Overexpress - NCI">{{cite web|title=overexpress|url=https://www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=45812|website=NCI Dictionary of Cancer Terms|publisher=National Cancer Institute at the National Institutes of Health|access-date=18 May 2017|quote=overexpress<br />In biology, to make too many copies of a protein or other substance. Overexpression of certain proteins or other substances may play a role in cancer development.|date = 2011-02-02}}</ref>}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==P== |
|||
{{glossary}} |
|||
{{term|[[p53]]}} |
|||
{{defn|}} |
|||
{{term|[[pachynema]]}} |
|||
{{ghat|Also '''pachytene stage'''.}} |
|||
{{defn|In {{gli|meiosis}}, the third of five substages of {{gli|prophase|prophase I}}, following {{gli|zygonema}} and preceding {{gli|diplonema}}. During pachynema, the {{gli|synaptonemal complex}} facilitates {{gli|crossing over}} between the synapsed {{gli|homologous chromosomes}}, and the {{gli|centrosomes}} begin to move apart from each other.<ref name="DoG7"/>}} |
|||
{{term|[[palindromic sequence]]}}{{anchor|palindromic sequences|palindrome|palindromes}} |
|||
{{ghat|Also '''palindrome'''.}} |
|||
{{defn|A {{gli|nucleic acid sequence}} of a double-stranded {{gli|DNA}} or {{gli|RNA}} molecule in which the unidirectional sequence (e.g. 5' to 3') of {{gli|nucleotides}} on one strand matches the sequence in the same direction (e.g. 5' to 3') on the {{gli|complementary}} strand. In other words, a nucleotide sequence is said to be palindromic if it is equal to its own {{gli|reverse complement}}. Palindromic {{gli|motifs}} are common in most genomes and are capable of forming {{gli|stem-loop|hairpins}}.}} |
|||
{{term|[[paracentric inversion]]}} |
|||
{{defn|A {{gli|chromosomal inversion}} in which the inverted segment does not include the chromosome's {{gli|centromere}}. Contrast ''{{gli|pericentric inversion}}''.}} |
|||
{{term|[[paralog]]}}{{anchor|paralogs|paralogous}} |
|||
{{defn|One of a set of {{gli|genes}} (or, more generally, any DNA sequences showing {{gli|sequence homology|homology}}) which are directly related to each other via one or more genetic {{gli|duplication}} events; such genes or sequences are said to be ''paralogous''. Paralogs result from the duplication of a single sequence within a single {{gli|genome}} and then the subsequent divergence of the duplicated sequences by mutation and natural selection (either within the original genome, or, during [[speciation]], in different genomes). Contrast ''{{gli|ortholog}}''.}} |
|||
{{term|parasitic DNA}} |
|||
{{defn|See ''{{gli|selfish genetic element}}''.}} |
|||
{{term|partial dominance}} |
|||
{{defn|See ''{{gli|incomplete dominance}}''.}} |
|||
{{term|[[particulate inheritance]]}} |
|||
{{defn|One of the defining ideas of {{gli|Mendelian inheritance}}, which holds that {{gli|phenotypic traits}} are or can be {{gli|inherited}} via the passing of "{{gli|gene|discrete particles}}" from {{gli|generation}} to generation. These particles may not have detectable effects in every generation but nevertheless retain their ability to be {{gli|expressed}} in subsequent generations.}} |
|||
{{term|passenger}} |
|||
{{defn|A DNA fragment of interest designed to be {{gli|splicing|spliced}} into a 'vehicle' such as a {{gli|plasmid}} {{gli|vector}} and then {{gli|molecular cloning|cloned}}.<ref name="DoG7"/>}} |
|||
{{term|patroclinous}} |
|||
{{defn|(of an {{gli|offspring}}) Resembling the male parent, genotypically or phenotypically, more closely than the female parent.<ref name="DoG7"/> Contrast ''{{gli|matroclinous}}''.}} |
|||
{{term|PCR}} |
|||
{{defn|See ''{{gli|polymerase chain reaction}}''.}} |
|||
{{term|PCR product}} |
|||
{{defn|See ''{{gli|amplicon}}''.}} |
|||
{{term|[[pedigree chart]]}} |
|||
{{defn|}} |
|||
{{term|[[penetrance]]}} |
|||
{{defn|The proportion of individuals with a given {{gli|genotype}} who express the associated {{gli|phenotype}}, usually given as a percentage. Because of the many complex interactions that govern {{gli|gene expression}}, the same {{gli|allele}} may produce an observable phenotype in one individual but not in another. If less than 100% of the individuals in a population carrying the genotype of interest also express the associated phenotype, both the genotype and phenotype may be said to show ''incomplete penetrance''. Penetrance quantifies the probability that an allele will result in the expression of its associated phenotype in any form, i.e. to any extent that makes an individual {{gli|carrier}} different from individuals without the allele. Compare ''{{gli|expressivity}}''.}} |
|||
{{term|[[peptide]]}}{{anchor|peptides}} |
|||
{{defn|A short chain of {{gli|amino acid}} [[monomer]]s linked by covalent {{gli|peptide bonds}}. Peptides are the fundamental building blocks of longer {{gli|polypeptide}} chains and hence of {{gli|proteins}}.}} |
|||
{{term|[[peptide bond]]}}{{anchor|peptide bonds}} |
|||
{{defn|A covalent chemical bond between the {{gli|carboxyl group}} of one {{gli|amino acid}} and the {{gli|amino group}} of an adjacent amino acid in a {{gli|peptide}} chain, formed by a [[dehydration reaction]] catalyzed by {{gli|peptidyl transferase}}, an enzyme within the {{gli|ribosome}}, during {{gli|translation}}.}} |
|||
{{term|pericentric}} |
|||
{{defn|(of a gene or region of a chromosome) Positioned near to the {{gli|centromere}} of the {{gli|chromosome}}.}} |
|||
{{term|[[pericentric inversion]]}} |
|||
{{defn|A {{gli|chromosomal inversion}} in which the inverted segment includes the chromosome's {{gli|centromere}}. Contrast ''{{gli|paracentric inversion}}''.}} |
|||
{{term|pervasive transcription}} |
|||
{{defn|}} |
|||
{{term|[[pharmacogenomics]]}} |
|||
{{defn|The study of the role played by the {{gli|genome}} in the body's response to pharmaceutical drugs, combining the fields of [[pharmacology]] and {{gli|genomics}}.}} |
|||
{{term|[[phenome]]}} |
|||
{{defn|The complete set of {{gli|phenotypes}} that are or can be expressed by a {{gli|genome}}, cell, tissue, organism, or species; the sum of all of its manifest chemical, morphological, and behavioral characteristics or traits.}} |
|||
{{term|phenomic lag}} |
|||
{{defn|A delay in the {{gli|phenotypic}} {{gli|expression}} of a genetic {{gli|mutation}} owing to the time required for the manifestation of changes in the affected biochemical pathways.<ref name="Rieger"/>}} |
|||
{{term|[[phenotype]]}}{{anchor|phenotypes|phenotypic}} |
|||
{{defn|The composite of the observable morphological, physiological, and behavioral {{gli|traits}} of an [[organism]] that result from the {{gli|expression}} of the organism's {{gli|genotype}} as well as the influence of environmental factors and the interactions between the two.}} |
|||
{{term|phosphate backbone}}{{anchor|phosphodiester backbone|phosphate–sugar backbone|sugar–phosphate backbone|backbone}} |
|||
{{ghat|Also '''phosphodiester backbone''', '''sugar–phosphate backbone''', and '''phosphate–sugar backbone'''.}} |
|||
{{defn|The linear chain of alternating phosphate and sugar compounds that results from the linking of consecutive {{gli|nucleotides}} in the same strand of a nucleic acid molecule, and which serves as the structural framework of the nucleic acid. Each individual {{gli|strand}} is held together by a single backbone consisting of a repeating series of {{gli|phosphodiester bonds}} between the phosphate group of one nucleotide and the {{gli|ribose}} or {{gli|deoxyribose}} sugars of the adjacent nucleotides. These bonds are created by {{gli|ligases}} and broken by {{gli|deoxyribonucleases}}.}} |
|||
{{term|[[phosphodiester bond]]}}{{anchor|phosphodiester bonds|phosphate bond|phosphate bonds}} |
|||
{{defn|A pair of [[ester]] bonds linking a [[phosphate]] molecule with the two [[pentose]] rings of consecutive {{gli|nucleosides}} on the same strand of a {{gli|nucleic acid}}. Each phosphate forms a covalent bond with the {{gli|3' carbon}} of one pentose and the {{gli|5' carbon}} of the adjacent pentose; the repeated series of such bonds that holds together a long chain of nucleotides in {{gli|DNA}} and {{gli|RNA}} is known as the {{gli|phosphate backbone|phosphate or phosphodiester backbone}}.}} |
|||
{{term|[[phylogenetics]]}}{{anchor|phylogenetic}} |
|||
{{defn|The study of the evolutionary history of and relationships between individuals or groups of organisms, such as species or populations, through methods that evaluate similarities and differences between observed {{gli|heredity|heritable}} {{gli|traits}}, including [[morphology (biology)|morphological]] features and {{gli|nucleic acid sequence|DNA sequences}}. The graphical presentation of data from such analyses is known as a ''[[phylogeny]]'' or ''phylogenetic tree''.}} |
|||
{{term|piRNA}} |
|||
{{defn|See ''{{gli|piRNA|Piwi-interacting RNA}}''.}} |
|||
<span id="piRNA"></span>{{term|[[piRNA|Piwi-interacting RNA]]}} |
|||
{{defn|}} |
|||
{{term|[[plasmid]]}}{{anchor|plasmids}} |
|||
{{defn|Any small {{gli|DNA}} molecule that is physically separated from the larger body of {{gli|genomic DNA|chromosomal DNA}} and can {{gli|DNA replication|replicate}} independently. Plasmids are most commonly found as small, circular, {{gli|double-stranded DNA}} molecules in [[prokaryote]]s such as [[bacteria]], though they are also sometimes present in [[archaea]] and [[eukaryote]]s.}} |
|||
{{term|[[pleiotropy]]}}{{anchor|pleiotropic}} |
|||
{{ghat|Also '''polypheny'''.}} |
|||
{{defn|The phenomenon by which one {{gli|gene}} influences two or more seemingly unrelated {{gli|phenotypic traits}}, by any of several distinct but potentially overlapping mechanisms.}} |
|||
{{term|[[ploidy]]}} |
|||
{{defn|The number of complete sets of {{gli|chromosomes}} in a [[cell (biology)|cell]], and hence the number of possible {{gli|alleles}} present within the cell at any given {{gli|autosomal}} {{gli|locus}}.}} |
|||
{{term|plus-strand}} |
|||
{{defn|See ''{{gli|coding strand}}''.}} |
|||
{{term|[[point mutation]]}}{{anchor|point mutations}} |
|||
{{defn|A {{gli|mutation}} by which a single {{gli|nucleotide}} base is changed, {{gli|insertion|inserted}}, or {{gli|deletion|deleted}} from a {{gli|sequence}} of DNA or RNA.}} |
|||
{{term|poly(A) tail}} |
|||
{{defn|}} |
|||
{{term|[[polyadenylation]]}}{{anchor|polyadenylated}} |
|||
{{defn|The addition of a series of multiple {{gli|adenosine}} {{gli|ribonucleotides}}, known as a {{gli|poly(A) tail}}, to the {{gli|3'-end}} of a {{gli|primary transcript|primary RNA transcript}}, typically a {{gli|mRNA|messenger RNA}}. A class of {{gli|post-transcriptional modification}}, polyadenylation serves different purposes in different cell types and organisms. In eukaryotes, the addition of a poly(A) tail is an important step in the processing of a raw transcript into a mature mRNA, ready for export to the cytoplasm where {{gli|translation}} occurs; in many bacteria, polyadenylation has the opposite function, instead promoting the RNA's degradation.}} |
|||
{{term|[[polygene]]}} |
|||
{{defn|}} |
|||
{{term|[[polygenic trait]]}} |
|||
{{defn|Any {{gli|phenotypic}} trait which is under the direct control of more than one {{gli|gene}}. Polygenic traits are often {{gli|quantitative traits}}.}} |
|||
{{term|polylinker}} |
|||
{{defn|See ''{{gli|multiple cloning site}}''.}} |
|||
{{term|[[polymerase]]}}{{anchor|polymerases}} |
|||
{{defn|Any of a class of enzymes which catalyze the synthesis of [[polymer]]ic molecules, especially {{gli|nucleic acid}} polymers, typically by encouraging the {{gli|pairing}} of free {{gli|nucleotides}} to those of an existing {{gli|complementary}} {{gli|template}}. {{gli|DNA polymerases}} and {{gli|RNA polymerases}} are essential for {{gli|DNA replication}} and {{gli|transcription}}, respectively.}} |
|||
<span id="PCR"></span>{{term|[[polymerase chain reaction]] (PCR)}}{{anchor|polymerase chain reaction}} |
|||
{{defn|Any of a wide variety of molecular biology methods involving the rapid production of millions or billions of copies of a specific {{gli|DNA sequence}}, allowing scientists to selectively {{gli|amplification|amplify}} fragments of a very small sample to a quantity large enough to study in detail. In its simplest form, PCR generally involves the incubation of a target DNA sample of known or unknown sequence with a reaction mixture consisting of {{gli|oligonucleotide}} {{gli|primers}}, a heat-stable {{gli|DNA polymerase}}, and free {{gli|deoxyribonucleotide triphosphates}} (dNTPs), all of which are supplied in excess. This mixture is then alternately heated and cooled to pre-determined temperatures for pre-determined lengths of time according to a specified pattern which is repeated for many cycles, typically in a {{gli|thermal cycler}} which automatically controls the required temperature variations. In each cycle, the most basic of which asincludes {{gli|denaturation}}, {{gli|annealing}}, and {{gli|elongation}} phases; the copies synthesized in the previous cycle are used as {{gli|template strand|templates}} for synthesis in the next cycle, resulting in a [[chain reaction]] that causes the exponential growth of the total number of copies in the reaction mixture. Amplification by PCR has become a standard technique in virtually all molecular biology laboratories.}} |
|||
{{term|[[genetic polymorphism|polymorphism]]}}{{anchor|polymorphisms|polymorphic}} |
|||
{{defn|The regular and simultaneous occurrence in the same population of two or more {{gli|alleles}} (or {{gli|genotypes}}) at the same {{gli|locus}} at {{gli|allele frequency|frequencies}} that cannot be accounted for by recurrent mutation alone (generally at least 1%), implying that the multiple alleles are being stably inherited by members of the population.<ref name="Rieger"/>}} |
|||
{{term|[[polypeptide]]}}{{anchor|polypeptides}} |
|||
{{defn|A long, continuous, and unbranched [[polymer]]ic chain of {{gli|amino acid}} [[monomer]]s linked by covalent {{gli|peptide bonds}}, typically longer than a {{gli|peptide}}. {{gli|protein|Proteins}} generally consist of one or more polypeptides arranged in a biologically functional way.}} |
|||
{{term|polypheny}} |
|||
{{defn|See ''{{gli|pleiotropy}}''.}} |
|||
{{term|[[polyploid]]}}{{anchor|polyploidy|polyploids}} |
|||
{{defn|(of a [[cell (biology)|cell]] or organism) Having more than two {{gli|homologous chromosomes|homologous}} copies of each {{gli|chromosome}}. Polyploidy may occur as a normal condition of chromosomes in certain cells or even entire organisms, or it may result from abnormal cell division or a mutation causing the {{gli|chromosomal duplication|duplication}} of the entire chromosome set. Contrast ''{{gli|haploid}}'' and ''{{gli|diploid}}''; see also ''{{gli|ploidy}}''.}} |
|||
{{term|[[polysome]]}} |
|||
{{ghat|Also '''polyribosome''' or '''ergosome'''.}} |
|||
{{defn|A complex of a {{gli|mRNA|messenger RNA}} molecule and two or more {{gli|ribosomes}} which act to {{gli|translate}} the mRNA transcript into a {{gli|polypeptide}}.}} |
|||
{{term|[[polysomy]]}} |
|||
{{defn|The condition of a cell or organism having at least one more copy of a particular {{gli|chromosome}} than is normal for its {{gli|ploidy}} level, e.g. a {{gli|diploid}} organism with three copies of a given chromosome is said to show {{gli|trisomy}}. Every polysomy is a type of {{gli|aneuploidy}}.}} |
|||
{{term|[[population genetics]]}} |
|||
{{defn|A subfield of {{gli|genetics}} and [[evolutionary biology]] that studies genetic differences within and between [[population]]s of organisms.}} |
|||
{{term|[[position effect]]}}{{anchor|position effects}} |
|||
{{defn|Any effect on the {{gli|expression}} or functionality of a {{gli|gene}} or sequence that is a consequence of its location or position within a {{gli|chromosome}} or other DNA molecule. A sequence's precise location relative to other sequences and structures tends to strongly influence its activity and other properties, because different {{gli|loci}} on the same molecule can have substantially different {{gli|genetic backgrounds}} and physical/chemical environments, which may also change over time. For example, the {{gli|transcription}} of a gene located very close to a {{gli|nucleosome}}, {{gli|centromere}}, or {{gli|telomere}} is often {{gli|repression|repressed}} or entirely prevented because the proteins that make up these structures block access to the DNA by {{gli|transcription factors}}, while the same gene is transcribed at a much higher rate when located in {{gli|euchromatin}}. Proximity to {{gli|promoters}}, {{gli|enhancers}}, and other {{gli|regulatory elements}}, as well as to regions of frequent {{gli|transposition}} by {{gli|mobile genetic element|mobile elements}}, can also directly affect expression; being located near the end of a chromosomal arm or to common {{gli|crossover}} points may affect when {{gli|DNA replication|replication}} occurs and the likelihood of {{gli|genetic recombination|recombination}}. Position effects are a major focus of research in the field of {{gli|epigenetic inheritance}}.}} |
|||
{{term|[[positional cloning]]}} |
|||
{{ghat|Also '''map-based cloning'''.}} |
|||
{{defn|A strategy for identifying and {{gli|molecular cloning|cloning}} a {{gli|candidate gene}} based on knowledge of its {{gli|locus}} or position alone and with little or no information about its {{gli|products}} or function, in contrast to {{gli|functional cloning}}. This method usually begins by comparing the {{gli|genomes}} of individuals expressing a {{gli|phenotype}} of unknown provenance (often a {{gli|genetic disorder|hereditary disease}}) and identifying {{gli|genetic markers}} shared between them. Regions defined by markers flanking one or more {{gli|genes of interest}} are cloned, and the genes located between the markers can then be identified by any of a variety of means, e.g. by {{gli|sequencing}} the region and looking for {{gli|open reading frames}}, by comparing the sequence and expression patterns of the region in {{gli|mutant}} and {{gli|wild-type}} individuals, or by testing the ability of the putative gene to {{gli|rescue}} a mutant phenotype.<ref name="DoG7"/>}} |
|||
{{term|positive (+) sense strand}} |
|||
{{defn|See ''{{gli|coding strand}}''.}} |
|||
{{term|[[post-transcriptional modification]]}}{{anchor|post-transcriptional modifications|post-transcriptional}} |
|||
{{defn|}} |
|||
{{term|[[post-translational modification]]}}{{anchor|post-translational modifications|post-translational}} |
|||
{{defn|}} |
|||
{{term|[[primary transcript]]}} |
|||
{{defn|The unprocessed, single-stranded {{gli|RNA}} molecule produced by the {{gli|transcription}} of a {{gli|DNA}} sequence as it exists before {{gli|post-transcriptional modifications}} such as {{gli|alternative splicing}} convert it into a mature RNA product such as an {{gli|mRNA}}, {{gli|tRNA}}, or {{gli|rRNA}}. A ''precursor mRNA'' or ''pre-mRNA'', for example, is a type of primary transcript that becomes a mature mRNA ready for {{gli|translation}} after processing.}} |
|||
{{term|[[primer (molecular biology)|primer]]}}{{anchor|primers}} |
|||
{{defn|A short, {{gli|single-stranded}} {{gli|oligonucleotide}}, typically 5–100 bases in length, which "primes" or initiates nucleic acid synthesis by {{gli|hybridization|hybridizing}} to a complementary sequence on a {{gli|template strand}} and thereby provides an existing {{gli|3'-end}} from which a {{gli|polymerase}} can {{gli|elongation|extend}} the new strand. Natural systems exclusively use {{gli|RNA}} primers to initiate {{gli|DNA replication}} and {{gli|transcription}}, whereas the ''{{gli|in vitro}}'' syntheses performed in many laboratory techniques such as {{gli|PCR}} often use {{gli|DNA}} primers. In modern laboratories, primers are carefully designed, often in "forward" and "reverse" pairs, to complement specific and unique sequences within {{gli|genomic DNA}}, with consideration given to their {{gli|melting}} and {{gli|annealing}} temperatures, and then purchased from commercial suppliers which create oligonucleotides on demand by ''de novo'' synthesis.}} |
|||
<span id="primer dimer"></span>{{term|[[primer dimer]] (PD)}} |
|||
{{defn|}} |
|||
{{term|priming}} |
|||
{{defn|The initiation of {{gli|nucleic acid}} synthesis by the {{gli|hybridization}} or {{gli|annealing}} of one or more {{gli|primers}} to a complementary sequence within a {{gli|template strand}}.}} |
|||
{{term|[[proband]]}} |
|||
{{ghat|Also '''prosposito''' for a male subject and '''prosposita''' for a female subject.}} |
|||
{{defn|A term used in {{gli|medical genetics}} and {{gli|genealogy}} to denote a particular subject being studied or reported on.}} |
|||
{{term|probe}}{{anchor|probes}} |
|||
{{defn|Any [[reagent]] used to make a single measurement in a biochemical assay such as a gene expression experiment. Compare ''{{gli|reporter}}''.}} |
|||
{{term|probe-set}} |
|||
{{defn|A collection of two or more {{gli|probe|probes}} designed to measure a single molecular species, such as a collection of {{gli|oligonucleotides}} designed to {{gli|hybridization|hybridize}} to various parts of the {{gli|mRNA}} transcripts generated from a single gene.}} |
|||
{{term|[[process molecular gene concept]]}} |
|||
{{defn|}} |
|||
{{term|[[prometaphase]]}} |
|||
{{defn|The second stage of [[cell division]] in {{gli|mitosis}}, following {{gli|prophase}} and preceding {{gli|metaphase}}, during which the nuclear membrane disintegrates, the chromosomes inside form {{gli|kinetochores}} around their {{gli|centromeres}}, [[microtubules]] emerging from the poles of the {{gli|mitotic spindle}} reach the nuclear space and attach to the kinetochores, and [[motor protein]]s associated with the microtubules begin to push the chromosomes toward the center of the cell.}} |
|||
{{term|[[promoter (genetics)|promoter]]}}{{anchor|promoters}} |
|||
{{defn|A sequence or region of DNA, usually 100–1,000 base pairs long, to which {{gli|transcription factors}} bind in order to recruit {{gli|RNA polymerase}} to the sequence and initiate the {{gli|transcription}} of one or more genes. Promoters are located {{gli|upstream}} of the genes they transcribe, near the transcription start site.}} |
|||
{{term|promotion}} |
|||
{{defn|See ''{{gli|upregulation}}''.}} |
|||
{{term|[[prophase]]}} |
|||
{{defn|The first stage of [[cell division]] in both {{gli|mitosis}} and {{gli|meiosis}}, occurring after {{gli|interphase}} and before {{gli|prometaphase}}, during which the DNA of the chromosomes is {{gli|DNA condensation|condensed}} into {{gli|chromatin}}, the {{gli|nucleolus}} disintegrates, {{gli|centrosomes}} move to opposite ends of the cell, and the {{gli|mitotic spindle}} forms.}} |
|||
{{term|[[protein]]}}{{anchor|proteins}} |
|||
{{defn|A linear [[polymer]]ic [[macromolecule]] composed of a series of {{gli|amino acids}} linked by {{gli|peptide bonds}}. Proteins carry out the majority of the [[chemical reaction]]s that occur inside living [[cell (biology)|cells]].}} |
|||
{{term|[[proteome]]}} |
|||
{{defn|The entire set of {{gli|proteins}} that is or can be {{gli|expressed}} by a particular {{gli|genome}}, cell, tissue, or species at a particular time (such as during a single lifespan or during a specific developmental stage) or under particular conditions (such as when compromised by a certain disease).}} |
|||
{{term|[[pseudoallele]]}} |
|||
{{defn|Any of two or more different genes or sequences which have the same or similar contributions to {{gli|phenotype}}, and thus appear to be genuine {{gli|alleles}}, but are not actually structurally allelic (i.e. they do not occupy homologous {{gli|locus|loci}} on {{gli|homologous chromosomes}}).<ref name="Lincoln">{{cite book|title=A Dictionary of Ecology, Evolution, and Systematics|author-first=Roger J.|author-last=Lincoln|date=1982|publisher=Cambridge University Press|location=New York|isbn=9780521239578}}</ref>}} |
|||
{{term|[[pseudogene]]}} |
|||
{{defn|A non-functional sequence of {{gli|DNA}} that resembles a functional {{gli|gene}}. Pseudogenes are typically superfluous copies of functional genes which have been {{gli|duplication|duplicated}} by natural processes, except that they lack {{gli|gene regulation|regulatory}} sequences necessary for proper {{gli|transcription}} or {{gli|translation}} or contain other defects such as {{gli|frameshift mutation|frameshift mutations}}, premature {{gli|stop codon|stop codons}}, or missing {{gli|intron|introns}}.}} |
|||
{{term|pseudopolyploidy}} |
|||
{{defn|no=1|The condition in which the number of {{gli|chromosomes}} in a chromosome set is {{gli|chromosome duplication|doubled}} (or tripled, etc.) but without a corresponding increase in the actual amount of genetic material (i.e. the {{gli|ploidy|ploidy level}}). This occurs when the chromosomes of a normal chromosome complement (e.g. {{gli|diploid}}) become fragmented into smaller pieces, increasing the total number of individual chromosomes but not creating additional homologous copies of those chromosomes (such that the cell remains diploid).<ref name="Lincoln"/>}} |
|||
{{defn|no=2|Any numerical relationship between chromosome sets in groups of related organisms which suggests that some of those organisms are {{gli|polyploid|polyploids}} of others when in fact they are not.<ref name="Lincoln"/>}} |
|||
{{term|[[Punnett square]]}} |
|||
{{defn|A tabular diagram used to predict the possible {{gli|genotypes}} that can be inherited by the {{gli|offspring}} of a particular {{gli|cross}} or breeding experiment by summarizing all of the various combinations of maternal {{gli|alleles}} with paternal alleles. The resulting table can then be used to determine the probabilities that the offspring will have a particular genotype. The usefulness of Punnett squares is limited to discrete {{gli|phenotype|phenotypes}} inherited according to simple {{gli|Mendelian inheritance|Mendelian}} patterns.}} |
|||
{{term|[[purebred]]}} |
|||
{{ghat|Also '''purebreed'''.}} |
|||
{{defn|}} |
|||
{{term|[[purine]]}}{{anchor|purines}} |
|||
{{ghat|Abbreviated in shorthand with the letter '''''{{font|R|font=courier|size=big}}'''''.}} |
|||
{{defn|A double-ringed [[heterocyclic]] organic compound which, along with {{gli|pyrimidine}}, is one of two molecules from which all {{gli|nitrogenous bases}} (including the {{gli|nucleobases}} used in {{gli|DNA}} and {{gli|RNA}}) are derived. {{gli|adenine|Adenine}} ({{font|A|font=courier|size=big}}) and {{gli|guanine}} ({{font|G|font=courier|size=big}}) are classified as purines. The letter {{font|R|font=courier|size=big}} is sometimes used to indicate a generic purine; e.g. in a nucleotide sequence read, {{font|R|font=courier|size=big}} may be used to indicate that either purine nucleobase, {{font|A|font=courier|size=big}} or {{font|G|font=courier|size=big}}, can be substituted at the indicated position.}} |
|||
{{term|putative gene}} |
|||
{{defn|A specific {{gli|nucleotide sequence}} suspected to be a functional {{gli|gene}} based on the identification of its {{gli|open reading frame}}. The gene is said to be "putative" in the sense that no function has yet been described for its {{gli|gene product|products}}.}} |
|||
{{term|[[pyrimidine]]}}{{anchor|pyrimidines}} |
|||
{{ghat|Abbreviated in shorthand with the letter '''''{{font|Y|font=courier|size=big}}'''''.}} |
|||
{{defn|A single-ringed [[heterocyclic]] organic compound which, along with {{gli|purine}}, is one of two molecules from which all {{gli|nitrogenous bases}} (including the {{gli|nucleobases}} used in {{gli|DNA}} and {{gli|RNA}}) are derived. {{gli|cytosine|Cytosine}} ({{font|C|font=courier|size=big}}), {{gli|thymine}} ({{font|T|font=courier|size=big}}), and {{gli|uracil}} ({{font|U|font=courier|size=big}}) are classified as pyrimidines. The letter {{font|Y|font=courier|size=big}} is sometimes used to indicate a generic pyrimidine; e.g. in a nucleotide sequence read, {{font|Y|font=courier|size=big}} may be used to indicate that either pyrimidine nucleobase – {{font|C|font=courier|size=big}}, {{font|T|font=courier|size=big}}, or {{font|U|font=courier|size=big}} – can be substituted at the indicated position.}} |
|||
{{term|[[pyrimidine dimer]]}} |
|||
{{defn|A type of {{gli|molecular lesion}} caused by photochemical damage to {{gli|DNA}} or {{gli|RNA}}, whereby exposure to [[ultraviolet]] (UV) radiation induces the formation of [[covalent bond]]s between {{gli|pyrimidine}} {{gli|nucleobase|bases}} occupying adjacent positions in the same polynucleotide {{gli|strand}}, which in turn may cause local conformational changes in {{gli|secondary structure}} and prevent {{gli|base pairing}} with the opposite strand. In DNA, the dimerization reaction occurs between neighboring {{gli|thymine}} and {{gli|cytosine}} residues ({{font|T|font=courier|size=big}}−{{font|T|font=courier|size=big}}, {{font|C|font=courier|size=big}}−{{font|C|font=courier|size=big}}, or {{font|T|font=courier|size=big}}−{{font|C|font=courier|size=big}}); it can also occur between cytosine and {{gli|uracil}} residues in {{gli|dsRNA|double-stranded RNA}}. Pyrimidine dimers are usually quickly corrected by {{gli|nucleotide excision repair}}, but uncorrected lesions can inhibit or arrest polymerase activity during {{gli|transcription}} or {{gli|DNA replication|replication}}.}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==Q== |
|||
{{glossary}} |
|||
{{term|[[quantitative genetics]]}} |
|||
{{defn|A branch of {{gli|population genetics}} which studies {{gli|phenotypes}} that vary continuously (such as height or mass) as opposed to those that fall into discretely identifiable categories (such as eye color or the presence or absence of a particular {{gli|trait}}). Quantitative genetics employs statistical methods and concepts to link continuously distributed phenotypic values to specific {{gli|genotypes}} and {{gli|gene products}}.}} |
|||
<span id="qPCR"></span>{{term|[[quantitative PCR]] (qPCR)}}{{anchor|quantitative PCR}} |
|||
{{ghat|Also '''real-time PCR (rtPCR)'''.}} |
|||
{{defn|}} |
|||
{{term|[[quantitative trait]]}}{{anchor|quantitative traits}} |
|||
{{ghat|Also '''complex trait'''.}} |
|||
{{defn|}} |
|||
<span id="QTL"></span>{{term|[[quantitative trait locus]] (QTL)}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
==R== |
|||
{{glossary}} |
|||
{{term|rDNA}} |
|||
{{defn|no=1|An abbreviation of {{gli|recombinant DNA}}.}} |
|||
{{defn|no=2|An abbreviation of {{gli|ribosomal DNA}}.}} |
|||
{{term|[[reading frame]]}} |
|||
{{defn|A way of dividing the {{gli|nucleotide sequence}} in a {{gli|DNA}} or {{gli|RNA}} molecule into a set of consecutive, non-overlapping triplets, which is "read" by {{gli|proteins}} during {{gli|transcription}} and {{gli|replication}}. In coding DNA, each triplet is referred to as a {{gli|codon}} that corresponds to a particular {{gli|amino acid}} during {{gli|translation}}. In general, only one reading frame (the so-called {{gli|open reading frame}}) in a given section of a nucleic acid can be used to make functional proteins, but there are exceptions in a few organisms. A {{gli|frameshift mutation}} results in a shift in the normal reading frame and affects all downstream codons.}} |
|||
{{term|real-time PCR (rtPCR)}} |
|||
{{defn|See ''{{gli|quantitative PCR}}''.}} |
|||
{{term|reassociation kinetics}} |
|||
{{defn|The measurement and manipulation of the rate of {{gli|annealing|reannealing}} of {{gli|complementary}} strands of {{gli|DNA}}, generally by heating and denaturing a {{gli|dsDNA|double-stranded}} molecule into {{gli|ssDNA|single strands}} and then observing their rehybridization at a cooler temperature. Because the {{gli|base pair}} {{font|G|font=courier|size=big}}+{{font|C|font=courier|size=big}} requires more energy to anneal than the base pair {{font|A|font=courier|size=big}}+{{font|T|font=courier|size=big}}, the rate of reannealing between two strands depends partly on their {{gli|nucleotide sequence}}, and it is therefore possible to predict or estimate the sequence of the duplex molecule by the time it takes to fully hybridize. Reassociation kinetics is studied with {{gli|C0t analysis}}: fragments reannealing at low C0t values tend to have highly {{gli|repetitive sequences}}, while higher C0t values imply more unique sequences.<ref name="DoG7"/>}} |
|||
{{term|[[dominance (genetics)|recessiveness]]}}{{anchor|recessive}} |
|||
{{defn|A relationship between the {{gli|alleles}} of a {{gli|gene}} in which one allele produces an effect on {{gli|phenotype}} that is overpowered or "masked" by the contribution of another allele at the same {{gli|locus}}; the first allele and its associated phenotypic {{gli|trait}} are said to be ''recessive'', and the second allele and its associated trait are said to be ''{{gli|dominance|dominant}}''. Often, recessive alleles code for inefficient or dysfunctional proteins. Like dominance, recessiveness is not an inherent property of any allele or phenotype, but simply describes its relationship to one or more other alleles or phenotypes. In genetics shorthand, recessive alleles are often represented by a lowercase letter (e.g. "a", in contrast to the dominant "A").}} |
|||
{{term|[[reciprocal cross]]}} |
|||
{{defn|A {{gli|crossbreeding}} experiment designed to test whether parental sex influences the inheritance of a particular trait. In crosses where the parents differ in {{gli|genotype}} or {{gli|phenotype}} or both, and hence only one of the parents (either the male or female parent) expresses the trait of interest, the reciprocal cross is the inverse, in which the parent of the other sex expresses the trait of interest instead. For example, if in the first cross a male expressing the trait is crossed with a female not expressing it, then in the reciprocal cross a female expressing the trait is crossed with a male not expressing it. By observing the {{gli|progeny}} resulting from each cross, geneticists can make inferences about which {{gli|sex chromosome}}, if either, influences the trait's expression.}} |
|||
{{term|reciprocal hybrid}} |
|||
{{defn|A {{gli|hybrid}} {{gli|offspring}} resulting from a {{gli|reciprocal cross}} between parents differing in genotype or phenotype or both.<ref name="DoG7"/>}} |
|||
{{term|[[chromosomal translocation|reciprocal translocation]]}} |
|||
{{defn|A type of {{gli|translocation|chromosomal translocation}} by which there is a reciprocal exchange of chromosome segments between two or more non-{{gli|homologous chromosomes|homologous}} {{gli|chromosomes}}. When the exchange of material is evenly balanced, reciprocal translocations are usually harmless.}} |
|||
[[File:Translocation-4-20.png|thumb|right|A '''{{gli|reciprocal translocation}}''' between chromosome 4 and chromosome 20]] |
|||
<span id="recombinant DNA"></span>{{term|[[recombinant DNA]] (rDNA)}} |
|||
{{defn|Any {{gli|DNA}} molecule in which laboratory methods of {{gli|genetic recombination}} have brought together genetic material from multiple sources, thereby creating a {{gli|nucleic acid sequence|sequence}} that would not otherwise be found in a naturally occurring {{gli|genome}}. Because DNA molecules from all organisms share the same basic chemical structure and properties, DNA sequences from any species, or even sequences created ''de novo'' by {{gli|artificial gene synthesis}}, may be incorporated into recombinant DNA molecules. Recombinant DNA technology is widely used in {{gli|genetic engineering}}.}} |
|||
{{term|[[recombinase]]}} |
|||
{{defn|}} |
|||
{{term|recombination}} |
|||
{{defn|See ''{{gli|genetic recombination}}'', ''{{gli|homologous recombination}}'', and ''{{gli|chromosomal crossover}}''.}} |
|||
{{term|recombinator}} |
|||
{{defn|Any {{gli|nucleotide sequence}} that increases the likelihood of {{gli|homologous recombination}} in nearby regions of the genome, e.g. the [[Chi sequence]] in certain species of bacteria.<ref name="DoG7"/>}} |
|||
{{term|recon}} |
|||
{{defn|The smallest unit of a DNA molecule capable of undergoing {{gli|homologous recombination}}, i.e. a pair of consecutive nucleotides, adjacent to each other in ''{{gli|cis}}''.<ref name="DoG7"/>}} |
|||
{{term|[[regulon]]}} |
|||
{{defn|A group of non-contiguous {{gli|genes}} which are {{gli|regulated}} as a unit, generally by virtue of having their {{gli|expression}} controlled by the same regulatory element or set of elements, e.g. the same {{gli|repressor}} or {{gli|activator}}. The term is most commonly used with prokaryotes, where a regulon may consist of genes from multiple {{gli|operons}}.}} |
|||
{{term|[[repeated sequence|repeat]]}}{{anchor|repeats|repeated sequence|repeated sequences|repeated|repeating}} |
|||
{{defn|}} |
|||
{{term|repetitive DNA}} |
|||
{{ghat|Also '''repetitious DNA'''.}} |
|||
{{defn|A region or fragment of DNA consisting largely or entirely of {{gli|repeated}} {{gli|nucleotide sequences}}.}} |
|||
{{term|replacement mutation}} |
|||
{{defn|See ''{{gli|nonsynonymous mutation}}''.}} |
|||
{{term|[[self-replication|replication]]}} |
|||
{{defn|no=1|The process by which certain biological molecules, notably the {{gli|nucleic acids}} {{gli|DNA}} and {{gli|RNA}}, produce copies of themselves.}} |
|||
{{defn|no=2|A technique used to estimate technical and biological variation in experiments for [[Multivariate analysis|statistical analysis]] of {{gli|DNA microarray|microarray}} data. Replicates may be ''technical replicates'', such as [[DNA microarray#Glossary|dye swaps]] or repeated array {{gli|hybridizations}}, or ''biological replicates'', biological samples from separate experiments which are used to test the effects of the same experimental treatment.}} |
|||
{{term|[[replication eye]]}} |
|||
{{ghat|Also '''replication bubble'''.}} |
|||
{{defn|The eye-shaped structure that forms when a pair of {{gli|replication forks}}, each growing away from the {{gli|origin of replication|origin}}, separates the strands of the double helix during {{gli|DNA replication}}.}} |
|||
{{term|[[replication fork]]}} |
|||
{{ghat|Also '''Y fork'''.}} |
|||
{{defn|}} |
|||
{{term|replication rate}} |
|||
{{defn|The speed at which {{gli|deoxyribonucleotides}} are incorporated into an elongating chain by {{gli|DNA polymerases}} during {{gli|DNA replication}}; or more generally the speed at which any {{gli|chromosome}}, genome, cell, or organism makes a complete, independently functional copy of itself.}} |
|||
{{term|replicator}} |
|||
{{defn|no=1|Any fragment or region of DNA that contains a {{gli|replication origin}}.<ref name="DoG7"/>}} |
|||
{{defn|no=2|Any molecule or structure capable of {{gli|replication|copying itself}}; namely, {{gli|nucleic acids}}, but also crystals of many minerals, e.g. [[kaolinite]].}} |
|||
{{term|[[replicon (genetics)|replicon]]}} |
|||
{{defn|Any molecule or region of {{gli|DNA}} or {{gli|RNA}} that {{gli|replication|replicates}} from a single {{gli|origin of replication}}.}} |
|||
{{term|[[replisome]]}} |
|||
{{defn|The entire complex of molecular machinery that carries out the process of {{gli|DNA replication}}, including all proteins, nucleic acids, and other molecules which participate at an active {{gli|replication fork}}.}} |
|||
{{term|reporter}} |
|||
{{defn|A {{gli|MIAME|MIAME-compliant}} term to describe a [[reagent]] used to make a single measurement in a gene expression experiment. MIAME defines it as "the nucleotide sequence present in a particular location on the array".<ref name="pmid18629115"/> A reporter may be a segment of single-stranded DNA that is [[covalent bond|covalently]] attached to the array surface. Compare ''{{gli|probe}}''.}} |
|||
{{term|[[reporter gene]]}} |
|||
{{defn|}} |
|||
{{term|repression}} |
|||
{{defn|See ''{{gli|downregulation}}''.}} |
|||
{{term|[[repressor]]}} |
|||
{{defn|A {{gli|DNA-binding protein}} that inhibits the {{gli|expression}} of one or more {{gli|genes}} by binding to the {{gli|operator}} and blocking the attachment of {{gli|RNA polymerase}} to the {{gli|promoter}}, thus preventing {{gli|transcription}}. This process is known as {{gli|negative control|negative gene regulation}}.}} |
|||
{{term|rescue}}{{anchor|rescued|rescues}} |
|||
{{defn|The restoration of a defective cell or tissue to a healthy or normal condition,<ref name="DoG7"/> or the {{gli|reversion}} or recovery of a mutant gene to its normal functionality, especially in the context of experimental genetics, where an experiment (e.g. a drug, {{gli|cross}}, or gene transfer) resulting in such a restoration is said to ''rescue'' the normal {{gli|phenotype}}.}} |
|||
{{term|residue}}{{anchor|residues}} |
|||
{{defn|An individual [[monomer]] or subunit of a larger [[polymer]]ic macromolecule; e.g. a {{gli|nucleic acid}} is composed of {{gli|nucleotide}} residues, and a {{gli|peptide}} or {{gli|protein}} is composed of {{gli|amino acid}} residues.<ref name="DoG7"/>}} |
|||
{{term|[[response element]]}} |
|||
{{defn|A short sequence of {{gli|DNA}} within a {{gli|promoter}} region that is able to bind specific {{gli|transcription factor|transcription factors}} in order to {{gli|gene regulation|regulate}} {{gli|transcription}} of specific genes.}} |
|||
{{term|restitution}} |
|||
{{defn|The spontaneous rejoining of an experimentally broken {{gli|chromosome}} which restores the original configuration.}} |
|||
{{term|restitution nucleus}} |
|||
{{defn|A {{gli|nucleus}} containing twice the expected number of chromosomes owing to an error in cell division, especially an unreduced, {{gli|diploid}} product of {{gli|meiosis}} resulting from the failure of the first or second meiotic division.}} |
|||
{{term|[[restriction enzyme]]}} |
|||
{{ghat|Also '''restriction endonuclease''', '''restriction exonuclease''', or '''restrictase'''.}}{{anchor|restriction endonuclease|restriction exonuclease|restrictase|restriction enzymes}} |
|||
{{defn|An {{gli|endonuclease}} or {{gli|exonuclease}} [[enzyme]] that recognizes and cleaves a nucleic acid molecule into {{gli|restriction fragment|fragments}} at or near specific recognition sequences known as {{gli|restriction site|restriction sites}} by breaking the {{gli|phosphodiester bond|phosphodiester bonds}} of the nucleic acid {{gli|phosphate backbone|backbone}}. Restriction enzymes are naturally occurring in many organisms, but are also routinely used for artificial modification of {{gli|DNA}} in laboratory techniques such as {{gli|molecular cloning}}.}} |
|||
{{term|[[restriction fragment]]}} |
|||
{{defn|Any {{gli|DNA}} fragment that results from the cutting of a DNA strand by a {{gli|restriction enzyme}} at one or more {{gli|restriction sites}}.}} |
|||
{{term|[[restriction fragment length polymorphism]] (RFLP)}} |
|||
{{defn|}} |
|||
{{term|[[restriction map]]}} |
|||
{{defn|A diagram of known {{gli|restriction sites}} within a known DNA sequence, such as a {{gli|plasmid}} {{gli|vector}}, obtained by systematically exposing the sequence to various {{gli|restriction enzymes}} and then comparing the lengths of the resulting {{gli|restriction fragment|fragments}}, a technique known as ''restriction mapping''. See also ''{{gli|gene map}}''.}} |
|||
{{term|[[restriction site]]}}{{anchor|restriction sites}} |
|||
{{ghat|Also '''restriction recognition site'''.}} |
|||
{{defn|A short, specific {{gli|sequence}} of nucleotides (typically 4 to 8 bases in length) that is reliably recognized by a particular {{gli|restriction enzyme}}. Because restriction enzymes usually bind as [[homodimer]]s, restriction sites are generally {{gli|palindromic sequences}} spanning both strands of a {{gli|dsDNA|double-stranded DNA}} molecule. Restriction {{gli|endonucleases}} cleave the {{gli|phosphate backbone}} between two nucleotides within the recognized sequence itself, but other types of restriction enzymes make their cuts at one end of the sequence or at a nearby sequence.}} |
|||
{{term|[[reverse genetics]]}} |
|||
{{defn|}} |
|||
{{term|[[reverse mutation]]}} |
|||
{{ghat|Also '''reversion'''.}} |
|||
{{defn|Any {{gli|mutation}} in a gene or DNA sequence which restores or {{gli|rescue|rescues}} the original function or phenotype that was altered or destroyed by a previous mutation in the same sequence.<ref name="DoG7"/> Contrast ''{{gli|forward mutation}}''; see also ''{{gli|suppressor mutation}}''.}} |
|||
{{term|[[reverse transcriptase]] (RT)}}{{anchor|reverse transcriptase}} |
|||
{{defn|An enzyme capable of synthesizing a {{gli|cDNA|complementary DNA}} molecule from an {{gli|RNA}} template, a process termed {{gli|reverse transcription}}.}} |
|||
{{term|[[reverse transcription]]}} |
|||
{{defn|The synthesis of a |
|||
{{gli|DNA}} molecule from an {{gli|RNA}} {{gli|template}}, the opposite of ordinary {{gli|transcription}}. This process, mediated by the enzyme {{gli|reverse transcriptase}}, is used by many viruses to {{gli|replicate}} their genomes, as well as by {{gli|retrotransposons}} and in eukaryotic cells.}} |
|||
{{term|[[revertant]]}} |
|||
{{defn|A gene or allele in which a {{gli|reverse mutation}} occurs, or an organism bearing such a gene or allele.<ref name="DoG7"/>}} |
|||
<span id="RNase"></span>{{term|[[ribonuclease]] (RNase)}}{{anchor|ribonucleases}} |
|||
{{defn|Any of a class of {{gli|nuclease}} enzymes which catalyze the [[hydrolytic]] cleavage of {{gli|phosphodiester bonds}} in {{gli|RNA}} molecules, thus severing {{gli|strands}} of {{gli|ribonucleotides}} and causing the degradation of RNA polymers into smaller components. Compare ''{{gli|deoxyribonuclease}}''.}} |
|||
<span id="rNA"></span>{{term|[[ribonucleic acid]] (RNA)}} |
|||
{{defn|A [[polymer]]ic {{gli|nucleic acid}} molecule composed of a series of {{gli|ribonucleotides}} which incorporate a set of four {{gli|nucleobase|nucleobases}}: {{gli|adenine}} ({{font|A|font=courier|size=big}}), {{gli|guanine}} ({{font|G|font=courier|size=big}}), {{gli|cytosine}} ({{font|C|font=courier|size=big}}), and {{gli|uracil}} ({{font|U|font=courier|size=big}}). Unlike {{gli|DNA}}, RNA is more often found as a {{gli|ssRNA|single strand}} folded onto itself, rather than a paired double strand. Various types of RNA molecules serve in a wide variety of essential biological roles, including {{gli|genetic code|coding}}, {{gli|translation|decoding}}, {{gli|gene regulation|regulating}}, and {{gli|expressing}} {{gli|genes}}, as well as functioning as signaling molecules and, in certain [[virus|viral]] genomes, as the primary genetic material itself.}} |
|||
{{term|[[ribonucleotide]]}}{{anchor|ribonucleotides}} |
|||
{{defn|A {{gli|nucleotide}} containing {{gli|ribose}} as its pentose sugar component, and the monomeric subunit of {{gli|ribonucleic acid}} (RNA) molecules. Ribonucleotides canonically incorporate any of four {{gli|nitrogenous bases}}: {{gli|adenine}} ({{font|A|font=courier|size=big}}), {{gli|guanine}} ({{font|G|font=courier|size=big}}), {{gli|cytosine}} ({{font|C|font=courier|size=big}}), and {{gli|uracil}} ({{font|U|font=courier|size=big}}). Compare ''{{gli|deoxyribonucleotide}}''.}} |
|||
<span id="ribonucleotide reductase"></span>{{term|[[ribonucleotide reductase]] (RNR)}} |
|||
{{ghat|Also '''ribonucleoside diphosphate reductase'''.}} |
|||
{{defn|An enzyme which catalyzes the formation of {{gli|deoxyribonucleotides}} via the [[reduction (chemistry)|reductive]] [[dehydroxylation]] of {{gli|ribonucleotides}}, specifically by removing the 2' hydroxyl group from the {{gli|ribose}} ring of {{gli|nucleoside diphosphate|ribonucleoside diphosphates}} (rNDPs). RNR plays a critical role in regulating the overall rate of DNA synthesis such that the ratio of DNA to cell mass is kept constant during cell division and {{gli|DNA repair}}.}} |
|||
<span id="ribonucleoprotein"></span>{{term|[[ribonucleoprotein]] (RNP)}} |
|||
{{defn|A {{gli|nucleoprotein}} that is a complex of one or more {{gli|RNA}} molecules and one or more {{gli|proteins}}. Examples include {{gli|ribosomes}} and the enzyme [[ribonuclease P]].}} |
|||
{{term|[[ribose]]}} |
|||
{{defn|A [[monosaccharide]] sugar which, as D-ribose in its [[pentose]] ring form, is one of three primary components of the {{gli|ribonucleotides}} from which {{gli|RNA|ribonucleic acid}} (RNA) molecules are built. Ribose differs from its structural analog {{gli|deoxyribose}}, used in {{gli|DNA}}, only at the 2' carbon, where ribose has an attached hydroxyl group that deoxyribose lacks.}} |
|||
<span id="ribosomal DNA"></span>{{term|[[ribosomal DNA]] (rDNA)}} |
|||
{{defn|}} |
|||
<span id="rRNA"></span>{{term|[[ribosomal RNA]] (rRNA)}} |
|||
{{defn|}} |
|||
{{term|[[ribosome]]}}{{anchor|ribosomes|ribosomal}} |
|||
{{defn|A molecular complex which serves as the site of {{gli|translation|protein synthesis}}. Ribosomes consist of two subunits (the ''small subunit'', which reads the messages encoded in {{gli|mRNA}} molecules, and the ''large subunit'', which links {{gli|amino acids}} in sequence to form a {{gli|polypeptide}} chain), each of which is composed of one or more strands of {{gli|rRNA|ribosomal RNA}} and various [[ribosomal protein]]s.}} |
|||
{{term|[[riboswitch]]}} |
|||
{{defn|A {{gli|regulatory}} sequence within a {{gli|messenger RNA}} {{gli|transcript}} that can bind a small effector molecule, preventing or disrupting {{gli|translation}} and thereby acting as a switch that regulates the mRNA's {{gli|expression}}.}} |
|||
{{term|[[ribozyme]]}} |
|||
{{defn|}} |
|||
{{term|RNA}} |
|||
{{defn|See ''{{gli|RNA|ribonucleic acid}}''.}} |
|||
<span id="rNA gene"></span>{{term|[[RNA gene]]}} |
|||
{{defn|A {{gli|gene}} that codes for any of the various types of {{gli|ncRNA|non-coding RNAs}} (e.g. {{gli|rRNA|rRNAs}} and {{gli|tRNA|tRNAs}}).<ref name="DoG7"/>}} |
|||
<span id="rNA interference"></span>{{term|[[RNA interference]] (RNAi)}} |
|||
{{defn|}} |
|||
<span id="rNA polymerase"></span>{{term|[[RNA polymerase]]}} |
|||
{{ghat|Often abbreviated '''RNAP''' or '''RNApol'''.}} |
|||
{{defn|Any of a class of {{gli|polymerase}} [[enzyme]]s that synthesize {{gli|RNA}} molecules from a {{gli|DNA}} template. RNA polymerases are essential for {{gli|transcription}} and are found in all living organisms and many viruses. They build long single-stranded polymers called {{gli|primary transcript|transcripts}} by adding {{gli|ribonucleotides}} one at a time in the {{gli|5'}}-to-{{gli|3'}} direction, relying on the {{gli|template strand|template}} provided by the {{gli|complementary}} strand to transcribe the nucleotide sequence faithfully.}} |
|||
<span id="rNA splicing"></span>{{term|[[RNA splicing]]}} |
|||
{{defn|}} |
|||
<span id="RISC"></span>{{term|[[RNA-induced silencing complex]] (RISC)}} |
|||
{{defn|A {{gli|ribonucleoprotein}} complex which works to {{gli|silencing|silence}} endogenous and exogenous genes by participating in various {{gli|RNAi|RNA interference}} pathways at the transcriptional and translational levels. RISC can bind both {{gli|ssRNA|single-stranded}} and {{gli|dsRNA|double-stranded RNA}} fragments and then cleave them or use them as guides to target complementary mRNAs for degradation.}} |
|||
{{term|RNase}} |
|||
{{defn|See ''{{gli|ribonuclease}}''.}} |
|||
<span id="Robertsonian translocation"></span>{{term|[[Robertsonian translocation]] (ROB)}} |
|||
{{defn|A type of {{gli|translocation|chromosomal translocation}} by which {{gli|double-strand breaks}} at or near the {{gli|centromeres}} of two {{gli|acrocentric}} {{gli|chromosomes}} cause a reciprocal exchange of segments that gives rise to one large {{gli|metacentric}} chromosome (composed of the {{gli|long arm|long arms}}) and one extremely small chromosome (composed of the {{gli|short arm|short arms}}), the latter of which is often subsequently lost from the cell with little effect because it contains very few genes. The resulting {{gli|karyotype}} shows one fewer than the expected total number of chromosomes, because two previously distinct chromosomes have essentially fused together. {{gli|carrier|Carriers}} of Robertsonian translocations are generally not associated with any phenotypic abnormalities, but do have an increased risk of generating meiotically unbalanced {{gli|gametes}}.}} |
|||
{{term|[[rolling circle replication]] (RCR)}} |
|||
{{defn|}} |
|||
{{term|rRNA}} |
|||
{{defn|See ''{{gli|rRNA|ribosomal RNA}}''.}} |
|||
{{term|rtPCR}} |
|||
{{defn|no=1|An abbreviation of ''real-time PCR'', synonymous with {{gli|quantitative PCR}}.}} |
|||
{{defn|no=2|An abbreviation of {{gli|reverse transcription PCR}}.}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==S== |
|||
{{glossary}} |
|||
{{term|samesense mutation}} |
|||
{{defn|See ''{{gli|synonymous mutation}}''.}} |
|||
{{term|[[Sanger sequencing]]}} |
|||
{{defn|A method of {{gli|DNA sequencing}} based on the ''in vitro'' {{gli|DNA replication|replication}} of a DNA {{gli|template strand|template}} sequence, during which [[fluorochrome]]-labeled, chain-terminating [[dideoxynucleotide]]s are randomly incorporated in the elongating strand; the resulting fragments are then sorted by size with {{gli|electrophoresis}}, and the particular fluorochrome terminating each of the size-sorted fragments is detected by laser chromatography, thus revealing the {{gli|sequence}} of the original DNA template through the order of the fluorochrome labels as one reads from small-sized fragments to large-sized fragments. Though Sanger sequencing has been replaced in some contexts by {{gli|next-generation sequencing|next-generation}} methods, it remains widely used for its ability to produce relatively long sequence reads (500+ {{gli|nucleotides}}) and its very low error rate.}} |
|||
[[File:Sanger-sequencing.svg|thumb|right|An outline of the '''{{gli|Sanger sequencing}}''' method]] |
|||
{{term|saturation hybridization}} |
|||
{{defn|An ''{{gli|in vitro}}'' nucleic acid {{gli|hybridization}} reaction in which one polynucleotide component (either {{gli|DNA}} or {{gli|RNA}}) is supplied in great excess relative to the other, causing all complementary sequences in the other polynucleotide to pair with the excess sequences and form hybrid {{gli|duplex}} molecules.<ref name="DoG7"/>}} |
|||
{{term|[[scaffolding (bioinformatics)|scaffolding]]}} |
|||
{{defn|}} |
|||
{{term|scRNA}} |
|||
{{defn|See ''{{gli|scRNA|small conditional RNA}}''.}} |
|||
{{term|[[selectable marker]]}} |
|||
{{defn|}} |
|||
{{term|[[selective sweep]]}} |
|||
{{defn|The process by which strong [[positive selection]] of a new and beneficial {{gli|mutation}} within a population causes the mutation to reach {{gli|fixation}} so quickly that nearby {{gli|linkage|linked}} DNA sequences also become fixed via {{gli|genetic hitchhiking}}, thereby reducing or eliminating the {{gli|genetic variation}} of nearby loci within the population.}} |
|||
{{term|[[selfish genetic element]]}} |
|||
{{ghat|Also '''selfish DNA''' or '''parasitic DNA'''.}} |
|||
{{defn|Any genetic material (e.g. a {{gli|gene}} or any other DNA sequence) which can enhance its own replication and/or transmission into subsequent generations at the expense of other genes in the genome, even if doing so has no positive effect or even a net negative effect on the fitness of the genome as a whole. Selfish elements usually work by producing self-acting {{gli|gene products}} which enable them to repeatedly {{gli|copy and paste}} themselves into other parts of the genome, independently of normal {{gli|DNA replication}} (as with {{gli|transposable elements}}); by facilitating the uneven swapping of chromosome segments during {{gli|genetic recombination}} events (as with {{gli|unequal crossing over}}); or by disrupting the normally equal redistribution of replicated material during {{gli|mitotis}} or {{gli|meiosis}} such that the probability that the selfish element is present in a given daughter cell is greater than the normal 50 percent (as with {{gli|gene drives}}).}} |
|||
{{term|[[semiconservative replication]]}} |
|||
{{defn|The standard mode of {{gli|DNA replication}} that occurs in all living cells, in which each of the two parental {{gli|strand|strands}} of the original {{gli|dsDNA|double-stranded DNA}} molecule are used as {{gli|template strand|template strands}}, with {{gli|DNA polymerase|DNA polymerases}} replicating each strand separately and simultaneously in {{gli|antiparallel}} directions. The result is that each of the two double-stranded daughter molecules is composed of one of the original parental strands and one newly synthesized complementary strand, such that each daughter molecule conserves the precise sequence of information (indeed the very same atoms) from one-half of the original molecule. Contrast ''{{gli|conservative replication}}'' and ''{{gli|dispersive replication}}''.}} |
|||
[[File:DNAreplicationModes.png|thumb|right|Three different modes of {{gli|DNA replication}}. In '''{{gli|semiconservative replication}}''', each of the two daughter molecules is built from one of the original parental strands and one newly synthesized strand. In '''{{gli|conservative replication}}''', the original parent molecule remains intact while the replicated molecule is composed of two newly synthesized strands. In '''{{gli|dispersive replication}}''', each of the daughter molecules is an uneven mix of old and new, with some segments consisting of the two parental strands and others consisting of two newly synthesized strands. Only semiconservative replication occurs naturally.]] |
|||
{{term|[[sense (molecular biology)|sense]]}} |
|||
{{defn|A distinction made between the individual {{gli|strand|strands}} of a {{gli|double-stranded DNA}} molecule in order to easily and specifically identify each strand. The two {{gli|complementarity|complementary}} strands are distinguished as ''sense'' and ''antisense'' or, equivalently, the ''{{gli|coding strand}}'' and the ''{{gli|template strand}}''. It is the antisense/template strand which is actually used as the template for {{gli|transcription}}; the sense/coding strand merely resembles the sequence of {{gli|codons}} on the RNA transcript, which makes it possible to determine from the DNA sequence alone the expected {{gli|amino acid}} sequence of any protein {{gli|translated}} from the RNA transcript. Which strand is which is relative only to a particular RNA transcript and not to the entire DNA molecule; that is, either strand can function as the sense/coding or antisense/template strand.}} |
|||
{{term|[[sense codon]]}} |
|||
{{defn|Any {{gli|codon}} that specifies an {{gli|amino acid}}, as opposed to a {{gli|stop codon}}, which does not specify any particular amino acid but instead signals the end of translation.}} |
|||
{{term|sequence}} |
|||
{{defn|See ''{{gli|nucleic acid sequence}}''.}} |
|||
<span id="sequence-tagged site"></span>{{term|[[sequence-tagged site]] (STS)}} |
|||
{{defn|Any DNA {{gli|nucleotide sequence|sequence}} that occurs exactly once within a particular {{gli|genome}}, and whose {{gli|locus|location}} and nucleotide sequence are known with confidence.}} |
|||
{{term|sex chromosome}} |
|||
{{defn|See ''{{gli|allosome}}''.}} |
|||
{{term|[[sex linkage]]}} |
|||
{{defn|}} |
|||
{{term|short arm}}{{anchor|short arms}} |
|||
{{ghat|Denoted in shorthand with the symbol '''''p'''''.}} |
|||
{{defn|In condensed {{gli|chromosomes}} where the positioning of the {{gli|centromere}} creates two segments or "arms" of unequal length, the shorter of the two arms of a {{gli|chromatid}}. Contrast ''{{gli|long arm}}''.}} |
|||
{{term|short tandem repeat (STR)}} |
|||
{{defn|See ''{{gli|microsatellite}}''.}} |
|||
<span id="SINE"></span>{{term|[[short interspersed nuclear element]] (SINE)}} |
|||
{{defn|}} |
|||
{{term|[[shotgun sequencing]]}} |
|||
{{defn|}} |
|||
{{term|[[silencer (genetics)|silencer]]}} |
|||
{{defn|A sequence or region of DNA that can be bound by a {{gli|repressor}}, thereby blocking the {{gli|transcription}} of a nearby gene.}} |
|||
{{term|silent allele}} |
|||
{{defn|An {{gli|allele}} that does not produce a detectable {{gli|gene product|product}}.<ref name="DoG7"/> Compare ''{{gli|null allele}}''.}} |
|||
{{term|[[silent mutation]]}} |
|||
{{defn|A type of {{gli|neutral mutation}} which does not have an observable effect on the organism's {{gli|phenotype}}. Though the term "silent mutation" is often used interchangeably with {{gli|synonymous mutation}}, synonymous mutations are not always silent, nor vice versa. {{gli|missense mutation|Missense mutations}} which result in a different {{gli|amino acid}} but one with similar functionality (e.g. [[leucine]] instead of [[isoleucine]]) are also often classified as silent, since such mutations usually do not significantly affect protein function.}} |
|||
{{term|simple sequence repeat (SSR)}} |
|||
{{defn|See ''{{gli|microsatellite}}''.}} |
|||
<span id="SNP"></span>{{term|[[single-nucleotide polymorphism]] (SNP)}} |
|||
{{defn|Any {{gli|substitution}} of a single {{gli|nucleotide}} which occurs at a specific position within a {{gli|genome}} and with measurable frequency within a population; for example, at a specific base position in a DNA sequence, the majority of the individuals in a population may have a {{gli|cytosine}} ({{font|C|font=courier|size=big}}), while in a minority of individuals, the same position may be occupied by an {{gli|adenine}} ({{font|A|font=courier|size=big}}). SNPs are usually defined with respect to a "standard" reference genome; an individual human genome differs from the reference human genome at an average of 4 to 5 million positions, most of which consist of SNPs and short {{gli|indel|indels}}. See also ''{{gli|polymorphism}}''.}} |
|||
<span id="single-strand break"></span>{{term|[[single-strand break]] (SSB)}} |
|||
{{defn|The loss of continuity of the {{gli|phosphate backbone|phosphate-sugar backbone}} in one {{gli|strand}} of a {{gli|double-stranded DNA|DNA duplex}}.<ref name="Rieger"/> See also ''{{gli|nick}}''; contrast ''{{gli|double-strand break}}''.}} |
|||
{{term|[[single-stranded]]}}{{anchor|single strand|single strands}} |
|||
{{defn|Composed of a single, unpaired {{gli|nucleic acid}} molecule, i.e. one linear {{gli|strand}} of {{gli|nucleotides}} sharing a single {{gli|phosphodiester backbone}}, as opposed to a {{gli|duplex}} of two such strands joined by base pairing. See also ''{{gli|ssDNA|single-stranded DNA}}'' and ''{{gli|ssRNA|single-stranded RNA}}''.}} |
|||
<span id="ssDNA"></span>{{term|[[single-stranded DNA]] (ssDNA)}} |
|||
{{defn|Any {{gli|DNA}} molecule that consists of a single nucleotide polymer, or "strand", as opposed to a pair of complementary strands held together by hydrogen bonds ({{gli|dsDNA|double-stranded DNA}}). In most circumstances, DNA is more stable and more common in double-stranded form, but high temperatures, low concentrations of dissolved salts, and very high or low pH can cause double-stranded molecules to decompose into two single-stranded molecules in a process known as "melting"; this reaction is exploited by naturally occurring enzymes such as those involved in {{gli|DNA replication}} as well as by laboratory techniques such as {{gli|polymerase chain reaction}}.}} |
|||
{{term|siRNA}} |
|||
{{defn|See ''{{gli|siRNA|small interfering RNA}}''.}} |
|||
{{term|[[sister chromatids]]}} |
|||
{{defn|A pair of identical copies ({{gli|chromatid|chromatids}}) produced as the result of the {{gli|DNA replication}} of a {{gli|chromosome}}, particularly when both copies are joined together by a common {{gli|centromere}}; the pair of sister chromatids is called a ''dyad''. The two sister chromatids are ultimately separated from each other into two different cells during {{gli|mitosis}} or {{gli|meiosis}}.}} |
|||
{{term|[[site-directed mutagenesis]]}} |
|||
{{defn|}} |
|||
<span id="scRNA"></span>{{term|[[small conditional RNA]] (scRNA)}} |
|||
{{defn|A class of small {{gli|RNA}} molecules engineered so as to change conformation conditionally in response to cognate molecular inputs, often with the goal of controlling signal transduction pathways ''{{gli|in vitro}}'' or ''{{gli|in vivo}}''.}} |
|||
<span id="siRNA"></span>{{term|[[small interfering RNA]] (siRNA)}} |
|||
{{defn|}} |
|||
<span id="snRNA"></span>{{term|[[small nuclear RNA]] (snRNA)}} |
|||
{{defn|}} |
|||
<span id="snoRNA"></span>{{term|[[small nucleolar RNA]] (snoRNA)}} |
|||
{{defn|}} |
|||
<span id="stRNA"></span>{{term|[[small temporal RNA]] (stRNA)}} |
|||
{{defn|A subclass of {{gli|miRNA|microRNAs}}, originally described in [[nematode]]s, which regulate the timing of developmental events by binding to complementary sequences in the {{gli|3' UTR|3' untranslated regions}} of {{gli|mRNA|messenger RNAs}} and inhibiting their {{gli|translation}}. In contrast to {{gli|siRNA|siRNAs}}, which serve similar purposes, stRNAs bind to their target mRNAs after the initiation of translation and without affecting mRNA stability, which makes it possible for the target mRNAs to resume translation at a later time.}} |
|||
{{term|snoRNA}} |
|||
{{defn|See ''{{gli|small nucleolar RNA}}''.}} |
|||
{{term|snRNA}} |
|||
{{defn|See ''{{gli|snRNA|small nuclear RNA}}''.}} |
|||
{{term|solenoid fiber}} |
|||
{{defn|}} |
|||
{{term|soluble RNA (sRNA)}} |
|||
{{defn|See ''{{gli|tRNA|transfer RNA}}''.}} |
|||
{{term|[[somatic cell]]}}{{anchor|somatic cells}} |
|||
{{ghat|Also '''vegetal cell''' or '''soma'''.}} |
|||
{{defn|Any biological [[cell (biology)|cell]] forming the body of an organism, or, in multicellular organisms, any cell other than a {{gli|gamete}}, {{gli|germ cell}}, or undifferentiated {{gli|stem cell}}. Somatic cells are theoretically distinct from cells of the {{gli|germ line}}, meaning the {{gli|mutations}} they have undergone can never be transmitted to the organism's descendants, though in practice exceptions do exist.}} |
|||
<span id="somatic cell nuclear transfer"></span>{{term|[[somatic cell nuclear transfer]] (SCNT)}} |
|||
{{defn|}} |
|||
{{term|somatic crossover}} |
|||
{{defn|See ''{{gli|mitotic recombination}}''.}} |
|||
{{term|[[Southern blot]]}} |
|||
{{defn|A {{gli|molecular biology}} method used for detecting a specific {{gli|nucleic acid sequence|sequence}} in {{gli|DNA}} samples. The technique combines separation of DNA fragments by {{gli|electrophoresis|gel electrophoresis}}, transfer of the DNA to a synthetic membrane, and subsequent identification of target fragments with radio-{{gli|labelling|labeled}} or fluorescent {{gli|hybridization probe|hybridization probes}}.}} |
|||
{{term|[[spacer DNA|spacer]]}} |
|||
{{ghat|Also '''intergenic spacer (IGS)''' or '''non-transcribed spacer (NTS)'''.}} |
|||
{{defn|Any sequence or region of {{gli|ncDNA|non-coding DNA}} separating neighboring {{gli|genes}}, whether {{gli|transcribed}} or not. The term is used in particular to refer to the non-coding regions between the many repeated copies of the {{gli|rRNA|ribosomal RNA}} genes.<ref name="Rieger"/> See also ''{{gli|intergenic region}}''.}} |
|||
{{term|spatially-restricted gene expression}} |
|||
{{defn|The {{gli|expression}} of one or more genes only within a specific anatomical region or tissue, often in response to a [[paracrine]] signal. The boundary between the jurisdictions of two spatially restricted genes may generate a sharp {{gli|phenotype|phenotypic}} gradient there, as with striping patterns.}} |
|||
<span id="spectral karyotype"></span>{{term|[[spectral karyotype]] (SKY)}} |
|||
{{defn|}} |
|||
{{term|[[spindle apparatus]]}}{{anchor|spindle|mitotic spindle|meiotic spindle}} |
|||
{{defn|}} |
|||
{{term|[[spliceosome]]}} |
|||
{{defn|}} |
|||
{{term|splicing}} |
|||
{{defn|See ''{{gli|genetic engineering}}''.}} |
|||
{{term|split-gene}} |
|||
{{defn|}} |
|||
{{term|ssDNA}} |
|||
{{defn|See ''{{gli|ssDNA|single-stranded DNA}}''.}} |
|||
{{term|ssRNA}} |
|||
{{defn|See ''{{gli|ssRNA|single-stranded RNA}}''.}} |
|||
<span id="SUT"></span>{{term|stable uncharacterized transcript (SUT)}} |
|||
{{defn|}} |
|||
{{term|standard genetic code}} |
|||
{{defn|The {{gli|genetic code}} used by the vast majority of living organisms for {{gli|translation|translating}} {{gli|nucleic acid sequence|nucleic acid sequences}} into {{gli|protein|proteins}}. In this system, of the 64 possible permutations of {{gli|codon|three-letter codons}} that can be made from the four {{gli|nucleotides}}, 61 code for one of the 20 {{gli|amino acids}}, and the remaining three code for {{gli|stop codon|stop signals}}. For example, the codon {{font|CAG|font=courier|size=big}} codes for the amino acid [[glutamine]] and the codon {{gli|ochre|{{font|UAA|font=courier|size=big}}}} is a stop codon. The standard genetic code is described as {{gli|degeneracy|degenerate}} or redundant because some amino acids can be coded for by more than one different codon.}} |
|||
[[File:GeneticCode21-version-2.svg|thumb|right|The '''{{gli|standard genetic code}}''' specifies a set of 20 different {{gli|amino acids}} from triplet arrangements of the four different {{gli|nucleobases}} used in {{gli|RNA}} polymers ({{gli|adenine|{{font|A|font=courier|size=big}}}}, {{gli|guanine|{{font|G|font=courier|size=big}}}}, {{gli|cytosine|{{font|C|font=courier|size=big}}}}, and {{gli|uracil|{{font|U|font=courier|size=big}}}}). To read this chart, choose one of the four letters in the innermost ring and then move outward, adding two more letters to complete a {{gli|codon}} triplet: a total of 64 unique codons can be made this way, 61 of which signal the addition of one of the 20 amino acids (identified by single-letter abbreviation as well as by full name and chemical structure) to a nascent {{gli|peptide}} chain, while the remaining three codons are {{gli|stop codon|stop codons}} signalling the termination of translation. The chart also indicates some of the chemical properties of the amino acids and the various ways in which they can be modified.]] |
|||
{{term|[[start codon]]}} |
|||
{{defn|The first {{gli|codon}} {{gli|translated}} by a {{gli|ribosome}} from a mature {{gli|mRNA|messenger RNA}} transcript, used as a signal to initiate {{gli|peptide}} synthesis. In the {{gli|standard genetic code}}, the start codon always codes for the same {{gli|amino acid}}, [[methionine]], in eukaryotes and for [[N-formylmethionine|a modified methionine]] in prokaryotes. The most common start codon is the triplet {{font|AUG|font=courier|size=big}}. Contrast ''{{gli|stop codon}}''.}} |
|||
{{term|[[statistical genetics]]}} |
|||
{{defn|A branch of genetics concerned with the development of [[statistics|statistical]] methods for drawing inferences from genetic data. The theories and methodologies of statistical genetics often support research in {{gli|quantitative genetics}}, {{gli|genetic epidemiology}}, and {{gli|bioinformatics}}.}} |
|||
{{term|[[stem-loop]]}} |
|||
{{ghat|Also '''hairpin''' or '''hairpin loop'''.}} |
|||
{{defn|}} |
|||
{{term|[[stem cell]]}} |
|||
{{defn|Any biological cell which has not yet [[cellular differentiation|differentiated]] into a specialized cell type and which can divide through {{gli|mitosis}} to produce more stem cells.}} |
|||
{{term|[[sticky end]]}} |
|||
{{defn|}} |
|||
{{term|[[stop codon]]}} |
|||
{{ghat|Also '''termination codon'''.}} |
|||
{{defn|A {{gli|codon}} that signals the termination of protein synthesis during {{gli|translation}} of a {{gli|mRNA|messenger RNA}} transcript. In the {{gli|standard genetic code}}, three different stop codons are used to dissociate {{gli|ribosome|ribosomes}} from the growing {{gli|amino acid}} chain, thereby ending translation: {{font|UAG|font=courier|size=big}} (nicknamed "amber"), {{font|UAA|font=courier|size=big}} ("ochre"), and {{font|UGA|font=courier|size=big}} ("opal"). Contrast ''{{gli|start codon}}''.}} |
|||
{{term|strand}}{{anchor|strands}} |
|||
{{defn|An individual chain of {{gli|nucleotides}} comprising a {{gli|nucleic acid}} polymer, existing either singly (in which case the nucleic acid molecule is said to be ''single-stranded'') or {{gli|paired}} in a {{gli|duplex}} (in which case it is said to be ''double-stranded'').}} |
|||
{{term|stringency}} |
|||
{{defn|The effect of conditions such as temperature and pH upon the degree of {{gli|complementarity}} that is required for a {{gli|hybridization}} reaction to occur between two single-stranded nucleic acid molecules. In the most stringent conditions, only exact complements can successfully hybridize; as stringency decreases, an increasing number of {{gli|mispairing|mismatches}} can be tolerated by the two hybridizing strands.<ref name="Lewin"/>}} |
|||
{{term|stRNA}} |
|||
{{defn|See ''{{gli|small temporal RNA}}''.}} |
|||
{{term|[[structural gene]]}}{{anchor|structural genes}} |
|||
{{defn|A {{gli|gene}} that codes for any protein or RNA {{gli|gene product|product}} other than a {{gli|gene regulation|regulatory factor}}. Structural gene products include [[enzyme]]s, structural proteins, and certain {{gli|non-coding RNAs}}.}} |
|||
{{term|[[submetacentric]]}} |
|||
{{defn|(of a linear {{gli|chromosome}} or chromosome fragment) Having a {{gli|centromere}} positioned close to but not exactly in the middle of the chromosome, resulting in {{gli|chromatid}} arms of slightly different lengths.<ref name="CoG"/> Compare ''{{gli|metacentric}}''.}} |
|||
{{term|[[base-pair substitution|substitution]]}} |
|||
{{defn|A type of {{gli|point mutation}} in which a single {{gli|nucleotide}} and its attached {{gli|nucleobase}} is replaced by a different nucleotide.}} |
|||
{{term|suppression}} |
|||
{{defn|See ''{{gli|downregulation}}''.}} |
|||
{{term|swivel point}} |
|||
{{defn|}} |
|||
{{term|[[synapsis]]}} |
|||
{{defn|}} |
|||
{{term|[[synaptonemal complex]]}} |
|||
{{defn|}} |
|||
{{term|syndesis}} |
|||
{{defn|The {{gli|synapsis}} of chromosomes during {{gli|meiosis}}.<ref name="DoG7"/>}} |
|||
{{term|synezis}} |
|||
{{defn|The aggregation of {{gli|chromosomes}} into a dense knot that adheres to one side of the {{gli|nucleus}}, commonly observed during {{gli|leptonema}} in certain organisms.<ref name="DoG7"/>}} |
|||
{{term|[[synonymous mutation]]}}{{anchor|synonymous mutations}} |
|||
{{ghat|Also '''synonymous substitution''' or '''samesense mutation'''.}} |
|||
{{defn|A type of {{gli|mutation}} in which the {{gli|substitution}} of one {{gli|nucleotide}} base for another results, after {{gli|transcription}} and {{gli|translation}}, in an amino acid sequence which is identical to the original unmutated sequence. This is possible because of the {{gli|degeneracy}} of the {{gli|genetic code}}, which allows different {{gli|codons}} to code for the same amino acid. Though synonymous mutations are often considered {{gli|silent mutation|silent}}, this is not always the case; a synonymous mutation may affect the efficiency or accuracy of {{gli|transcription}}, {{gli|RNA splicing|splicing}}, {{gli|translation}}, or any other process by which genes are {{gli|expressed}}, and thus become effectively non-silent. Contrast ''{{gli|nonsynonymous mutation}}''.}} |
|||
{{term|[[synteny]]}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==T== |
|||
{{glossary}} |
|||
{{term|[[tandem repeat]]}}{{anchor|tandem repeats}} |
|||
{{defn|A pattern within a {{gli|nucleic acid sequence}} in which one or more {{gli|nucleobases}} are repeated and the repetitions are directly adjacent (i.e. tandem) to each other. An example is {{font|ATGACATGACATGAC|font=courier|size=big}}, in which the sequence {{font|ATGAC|font=courier|size=big}} is repeated three times.}} |
|||
{{term|[[TATA box]]}} |
|||
{{ghat|Also '''Goldberg-Hogness box'''.}} |
|||
{{defn|A highly conserved {{gli|non-coding DNA}} sequence containing a {{gli|consensus sequence|consensus}} of repeating {{font|{{gli|thymine|T}}|font=courier|size=big}} and {{font|{{gli|adenine|A}}|font=courier|size=big}} base pairs that is commonly found in {{gli|promoter|promoter regions}} of genes in [[archaea]] and [[eukaryote]]s. The TATA box often serves as the site of initiation of {{gli|transcription}} or as a binding site for {{gli|transcription factor|transcription factors}}.}} |
|||
{{term|telestability}} |
|||
{{defn|Structural destabilization of the {{gli|DNA}} {{gli|double helix}} at a {{gli|locus}} that is relatively distant from the site of binding of a {{gli|DNA-binding protein}}.<ref name="DoG7"/>}} |
|||
{{term|[[centromere#telocentric|telocentric]]}} |
|||
{{defn|(of a linear {{gli|chromosome}} or chromosome fragment) Having a {{gli|centromere}} positioned at the terminal end of the chromosome (near or within the {{gli|telomere}}), resulting in only a single arm.<ref name="CoG"/> Compare ''{{gli|acrocentric}}''.}} |
|||
{{term|[[telomere]]}}{{anchor|telomeres}} |
|||
{{defn|A region of {{gli|repetitive DNA|repetitive}} {{gli|nucleic acid sequence|nucleotide sequences}} at each end of a linear {{gli|chromosome}} which protects the end of the chromosome from deterioration and from fusion with other chromosomes. Since each round of {{gli|DNA replication|replication}} results in the shortening of the chromosome, telomeres act as disposable buffers which are sacrificed to perpetual truncation instead of nearby genes; telomeres can also be lengthened by the enzyme [[telomerase]].}} |
|||
{{term|telomeric silencing}} |
|||
{{defn|The {{gli|repression}} of {{gli|transcription}} of genes in regions adjacent to {{gli|telomere|telomeres}}. Telomeres also appear to reduce the accessibility of subtelomeric {{gli|chromatin}} to modification by DNA {{gli|methyltransferase|methyltransferases}}.<ref name="DoG7"/>}} |
|||
{{term|[[telophase]]}} |
|||
{{defn|The final stage of [[cell division]] in both {{gli|mitosis}} and {{gli|meiosis}}, occurring after {{gli|anaphase}} and before or simultaneously with {{gli|cytokinesis}}, during which a nuclear membrane is synthesized around each set of {{gli|chromatids}}, {{gli|nucleolus|nucleoli}} are reassembled, and the {{gli|mitotic spindle}} is disassembled. Following cytokinesis, the new daughter cells resume {{gli|interphase}}.}} |
|||
{{term|[[template strand]]}}{{anchor|template}} |
|||
{{ghat|Also '''antisense strand''', '''negative (-) sense strand''', and '''noncoding strand'''.}} |
|||
{{defn|The {{gli|strand}} of a {{gli|dsDNA|double-stranded DNA}} molecule which is used as a template for RNA synthesis during {{gli|transcription}}. The sequence of the template strand is {{gli|complementary}} to the resulting RNA transcript. Contrast ''{{gli|coding strand}}''; see also ''{{gli|sense}}''.}} |
|||
{{term|terminalization}} |
|||
{{defn|In cytology, the progressive shift of {{gli|chiasmata}} from their original to more distal positions as {{gli|meiosis}} proceeds through {{gli|diplonema}} and {{gli|diakinesis}}.<ref name="DoG7"/>}} |
|||
{{term|termination codon}} |
|||
{{defn|See ''{{gli|stop codon}}''.}} |
|||
{{term|three-prime end}} |
|||
{{defn|See ''{{gli|3'-end}}''.}} |
|||
{{term|three-prime untranslated region}} |
|||
{{defn|See ''{{gli|3' untranslated region}}''.}} |
|||
<span id="thymidine"></span>{{term|term=[[thymidine]]|content=[[thymidine]] ({{font|T|font=courier|size=large}}, {{font|dT|font=courier|size=large}})}} |
|||
{{ghat|Also '''deoxythymidine'''.}} |
|||
{{defn|One of the four standard {{gli|nucleosides}} used in {{gli|DNA}} molecules, consisting of a {{gli|thymine}} {{gli|base}} with its N<sub>9</sub> nitrogen {{gli|glycosidic bond|bonded}} to the C<sub>1</sub> carbon of a {{gli|deoxyribose}} sugar. The prefix ''deoxy-'' is commonly omitted, since there are no ribonucleoside analogs of thymidine used in RNA, where it is replaced with {{gli|uridine}} instead.}} |
|||
<span id="thymine"></span>{{term|term=[[thymine]]|content=[[thymine]] ({{font|T|font=courier|size=large}})}} |
|||
{{ghat|Also '''5-methyluracil'''.}} |
|||
{{defn|A {{gli|pyrimidine}} {{gli|nucleobase}} used as one of the four standard nucleobases in {{gli|DNA}} molecules. Thymine forms a {{gli|base pair}} with {{gli|adenine}}. In {{gli|RNA}}, thymine is not used at all, and is instead replaced with {{gli|uracil}}.}} |
|||
{{term|thymine dimer}} |
|||
{{defn|See ''{{gli|pyrimidine dimer}}''.}} |
|||
{{term|tissue-specific gene expression}} |
|||
{{defn|Gene function and {{gli|expression}} which is restricted to a particular [[tissue (biology)|tissue]] or cell type. Tissue-specific expression is usually the result of an {{gli|enhancer}} which is activated only in the proper cell type.}} |
|||
{{term|[[topoisomerase]]}} |
|||
{{defn|Any of a class of {{gli|DNA-binding protein|DNA-binding}} enzymes which catalyze changes in the topological state of a {{gli|dsDNA|double-stranded DNA}} molecule by {{gli|nicking}} or {{gli|cutting}} the {{gli|sugar-phosphate backbone}} of one or both strands, relaxing the torsional stress inherent in the {{gli|double helix}} and unwinding or untangling the paired strands before {{gli|ligation|re-ligating}} the nicks. This process is usually necessary prior to {{gli|DNA replication|replication}} and {{gli|transcription}}. Topoisomerases thereby convert DNA between its {{gli|B-DNA|relaxed}} and {{gli|supercoiled}}, [[catenane|linked]] and unlinked, and [[molecular knot|knotted]] and unknotted forms without changing the sequence or overall chemical composition, such that the substrate and product molecules are structural isomers, differing only in their shape and their {{gli|twisting number|twisting}}, {{gli|linking number|linking}}, and/or {{gli|writhing number|writhing numbers}}.}} |
|||
{{term|trailer sequence}} |
|||
{{defn|See ''{{gli|3' untranslated region}}''.}} |
|||
{{term|trait}}{{anchor|traits|phenotypic trait|phenotypic traits}} |
|||
{{defn|}} |
|||
{{term|''trans''}} |
|||
{{defn|On the opposite side; across from; {{gli|trans-acting|acting}} from a different molecule. Contrast ''{{gli|cis}}''.}} |
|||
{{term|[[trans-acting|''trans''-acting]]}} |
|||
{{defn|Affecting a {{gli|gene}} or sequence on a different nucleic acid molecule or {{gli|strand}}. A {{gli|locus}} or sequence within a particular DNA molecule such as a {{gli|chromosome}} is said to be ''trans''-acting if it or its {{gli|gene product|products}} influence or act upon other sequences located relatively far away or on an entirely different molecule or chromosome. For example, a {{gli|DNA-binding protein}} acts "in ''trans''" if it binds to or interacts with a sequence located on any strand or molecule different from the one on which it is encoded. Contrast ''{{gli|cis-acting}}''.}} |
|||
{{term|[[trans-splicing|''trans''-splicing]]}} |
|||
{{defn|}} |
|||
{{term|transcribed spacer}} |
|||
{{defn|A {{gli|spacer}} sequence that is transcribed and thus included in the primary {{gli|rRNA|ribosomal RNA}} {{gli|transcript}} (as opposed to a {{gli|non-transcribed spacer}}) but subsequently excised and discarded during the maturation of functional RNAs of the {{gli|ribosome}}.<ref name="DoG7"/>}} |
|||
{{term|[[transcript (biology)|transcript]]}}{{anchor|transcripts}} |
|||
{{defn|A product of {{gli|transcription}}; that is, any {{gli|RNA}} molecule which has been synthesized by {{gli|RNA polymerase}} using a complementary {{gli|DNA}} molecule as a {{gli|template}}. When transcription is completed, transcripts separate from the DNA and become independent {{gli|primary transcripts}}. Particularly in eukaryotes, multiple {{gli|post-transcriptional modifications}} are usually necessary for raw transcripts to be converted into stable and persistent molecules, which are then described as ''mature'', though not all transcribed RNAs undergo maturation. Many transcripts are accidental, spurious, incomplete, or defective; others are able to perform their functions immediately and without modification, such as certain {{gli|ncRNA|non-coding RNAs}}.}} |
|||
{{term|[[transcript of unknown function]] (TUF)}} |
|||
{{defn|}} |
|||
{{term|transcriptase}} |
|||
{{defn|See ''{{gli|RNA polymerase}}''.}} |
|||
{{term|[[Transcription (biology)|transcription]]}}{{anchor|transcriptional|transcriptionally|transcribe|transcribes|transcribed}} |
|||
{{defn|The first step in the process of {{gli|gene expression}}, in which an {{gli|RNA}} molecule, known as a {{gli|transcript}}, is synthesized by [[enzyme]]s called {{gli|RNA polymerases}} using a {{gli|complementary}} {{gli|gene}} or other {{gli|DNA}} sequence as a {{gli|template}}. Transcription is a critical and fundamental process in all living organisms and is mandatory in order to make use of the information encoded within a {{gli|genome}}. All classes of RNA must be transcribed before they can exert their effects upon a cell. {{gli|mRNA|Messenger RNA}} (mRNA) in particular must proceed to {{gli|translation}} before a functional {{gli|protein}} can be produced, though the many types of {{gli|non-coding RNA}} fulfill their duties without being translated. Transcription is also not necessarily always beneficial for a cell: when the process occurs at the wrong time or at a {{gli|junk DNA|functionless locus}}, or when {{gli|mobile genetic element|mobile elements}} or infectious pathogens utilize their host's transcription machinery, the resulting transcripts (not to mention the waste of valuable energy and resources) can often be harmful to the host cell or genome.}} |
|||
<span id="transcription factor"></span>{{term|[[transcription factor]] (TF)}}{{anchor|transcription factors}} |
|||
{{defn|Any {{gli|protein}} that controls the rate of {{gli|transcription}} of genetic information from {{gli|DNA}} to {{gli|mRNA|messenger RNA}} by binding to a specific {{gli|nucleic acid sequence|DNA sequence}} and {{gli|activator|promoting}} or {{gli|repressor|blocking}} the recruitment of {{gli|RNA polymerase}} to nearby {{gli|genes}}. Transcription factors can effectively turn "on" and "off" specific genes in order to make sure they are {{gli|expressed}} at the right times and in the right places; for this reason, they are a fundamental and ubiquitous mechanism of {{gli|gene regulation}}.}} |
|||
{{term|transcription unit}} |
|||
{{defn|}} |
|||
{{term|[[transcriptional bursting]]}} |
|||
{{defn|The intermittent nature of {{gli|transcription}} and {{gli|translation}} mechanisms. Both processes occur in "bursts" or "pulses", with periods of gene activity separated by irregular intervals.}} |
|||
{{term|[[transcriptome]]}} |
|||
{{defn|The entire set of {{gli|RNA}} molecules (often referring to all types of RNA but sometimes exclusively to {{gli|mRNA|messenger RNA}}) that is or can be {{gli|expressed}} by a particular {{gli|genome}}, cell, population of cells, or species at a particular time or under particular conditions. The transcriptome is distinct from the {{gli|exome}} and the {{gli|translatome}}.}} |
|||
{{term|[[transduction (genetics)|transduction]]}} |
|||
{{defn|The transfer of genetic material between cells by a virus or viral vector, either naturally or artificially.}} |
|||
{{term|[[transfection]]}} |
|||
{{defn|The deliberate experimental introduction of exogenous {{gli|nucleic acids}} into a cell or embryo. In the broadest sense the term may refer to any such transfer and is sometimes used interchangeably with {{gli|transformation}}, though some applications restrict the usage of transfection to the introduction of naked or purified non-viral {{gli|DNA}} or {{gli|RNA}} into cultured eukaryotic cells (especially animal cells) resulting in the subsequent incorporation of the foreign DNA into the host {{gli|genome}} or the non-hereditary modification of {{gli|gene expression}} by the foreign RNA. As a contrast to both standard non-viral transformation and {{gli|transduction}}, transfection has also occasionally been used to refer to the uptake of purified viral nucleic acids by bacteria or plant cells without the aid of a viral vector.<ref name="DoG7"/>}} |
|||
<span id="tRNA"></span>{{term|[[transfer RNA]] (tRNA)}}{{anchor|transfer RNA|transfer RNAs|tRNAs}} |
|||
{{ghat|Formerly referred to as '''soluble RNA (sRNA)'''.}} |
|||
{{defn|A special class of {{gli|RNA}} molecule, typically 76 to 90 {{gli|nucleotides}} in length, that serves as a physical adapter allowing {{gli|mRNA}} transcripts to be {{gli|translated}} into sequences of {{gli|amino acids}} during protein synthesis. Each tRNA contains a specific {{gli|anticodon}} triplet corresponding to an amino acid that is covalently attached to the tRNA's opposite end; as translation proceeds, tRNAs are recruited to the {{gli|ribosome}}, where each mRNA {{gli|codon}} is paired with a tRNA containing the complementary anticodon. Depending on the organism, cells may employ as many as 41 distinct tRNAs with unique anticodons; because of {{gli|degeneracy|codon degeneracy}} within the {{gli|genetic code}}, several tRNAs containing different anticodons carry the same amino acid.}} |
|||
<span id="tmRNA"></span>{{term|[[transfer-messenger RNA]] (tmRNA)}} |
|||
{{defn|A type of RNA molecule in some bacteria which has dual {{gli|tRNA}}-like and {{gli|mRNA}}-like properties, allowing it to simultaneously perform a number of different functions during {{gli|translation}}.}} |
|||
{{term|tRNA-ligase}} |
|||
{{defn|See ''{{gli|aminoacyl-tRNA synthetase}}''.}} |
|||
{{term|[[transformant]]}}{{anchor|transformants}} |
|||
{{defn|A cell or organism which has taken up extracellular DNA by {{gli|transformation}} and which can express genes encoded by it.}} |
|||
{{term|[[transformation (genetics)|transformation]]}}{{anchor|transform|transformed|transforming|transformations}} |
|||
{{defn|}} |
|||
{{term|[[transgene]]}}{{anchor|transgenes|transgenic}} |
|||
{{defn|Any {{gli|gene}} or other segment of genetic material that has been isolated from one organism and then transferred either naturally or by any of a variety of {{gli|genetic engineering}} techniques into another organism, especially one of a different species. Transgenes are usually introduced into the second organism's {{gli|germ line}}. They are commonly used to study gene function or to confer an advantage not otherwise available in the unaltered organism.}} |
|||
{{term|[[transition (genetics)|transition]]}} |
|||
{{defn|A {{gli|point mutation}} in which a {{gli|purine}} nucleotide is substituted for another purine ({{font|{{gli|adenine|A}}|font=courier|size=big}} ↔ {{font|{{gli|guanine|G}}|font=courier|size=big}}) or a {{gli|pyrimidine}} nucleotide is substituted for another pyrimidine ({{font|{{gli|cytosine|C}}|font=courier|size=big}} ↔ {{font|{{gli|thymine|T}}|font=courier|size=big}}). Contrast ''{{gli|transversion}}''.}} |
|||
{{term|[[Translation (biology)|translation]]}}{{anchor|translational|translationally|translate|translates|translated}} |
|||
{{defn|The second step in the process of {{gli|gene expression}}, in which the {{gli|mRNA|messenger RNA}} transcript produced during {{gli|transcription}} is read by a {{gli|ribosome}} to produce a functional {{gli|protein}}.}} |
|||
{{term|[[translatome]]}} |
|||
{{defn|The entire set of {{gli|mRNA|messenger RNA}} molecules that are {{gli|translated}} by a particular {{gli|genome}}, cell, tissue, or species at a particular time or under particular conditions. Like the {{gli|transcriptome}}, it is often used as a proxy for quantifying levels of {{gli|gene expression}}, though the transcriptome also includes many RNA molecules that are never translated.}} |
|||
{{term|[[Chromosomal translocation|translocation]]}}{{anchor|translocations}} |
|||
{{defn|A type of [[chromosomal abnormality]] caused by the structural rearrangement of large sections of one or more {{gli|chromosomes}}. There are two main types: {{gli|reciprocal translocation|reciprocal}} and {{gli|Robertsonian translocation|Robertsonian}}.}} |
|||
{{term|transmission genetics}} |
|||
{{defn|The branch of genetics that studies the mechanisms involved in the transfer of genes from parents to offspring.<ref name="DoG7"/>}} |
|||
<span id="transposable element"></span>{{term|[[transposable element]] (TE)}}{{anchor|transposable elements|transposon|transposons}} |
|||
{{ghat|Also '''transposon'''.}} |
|||
{{defn|}} |
|||
{{term|[[transposase]]}} |
|||
{{defn|Any of a class of self-acting enzymes capable of {{gli|DNA-binding protein|binding}} to the flanking sequences of the {{gli|transposable element}} which encodes them and catalyzing its movement to another part of a genome, typically by an excision/insertion mechanism or a replicative mechanism, in a process known as {{gli|transposition}}.}} |
|||
{{term|[[transposition (genetics)|transposition]]}} |
|||
{{defn|The process by which a nucleic acid sequence known as a {{gli|transposable element}} changes its position within a {{gli|genome}}, either by excising and re-inserting itself at a different {{gli|locus}} (cut-and-paste) or by {{gli|duplicating}} itself without moving the element from its original locus (copy-paste). These reactions are catalyzed by an enzyme known as a {{gli|transposase}} which is encoded by a gene within the transposable element itself; thus the element's products are self-acting and can autonomously direct their own reproduction. Transposed sequences may re-insert at random loci or at sequence-specific targets, either on the same DNA molecule or on different molecules.}} |
|||
{{term|[[transvection (genetics)|transvection]]}} |
|||
{{defn|}} |
|||
{{term|[[transversion]]}} |
|||
{{defn|A {{gli|point mutation}} in which a {{gli|purine}} nucleotide is substituted for a {{gli|pyrimidine}} nucleotide, or vice versa (e.g. {{font|{{gli|adenine|A}}|font=courier|size=big}} ↔ {{font|{{gli|cytosine|C}}|font=courier|size=big}} or {{font|{{gli|adenine|A}}|font=courier|size=big}} ↔ {{font|{{gli|thymine|T}}|font=courier|size=big}}). Contrast ''{{gli|transition}}''.}} |
|||
{{term|[[trinucleotide repeat]]}} |
|||
{{defn|Any sequence in which an individual nucleotide {{gli|triplet}} is {{gli|repeated}} many times {{gli|tandem repeat|in tandem}}, whether in a gene or non-coding sequence. At most {{gli|loci}} some degree of repetition is normal and harmless, but mutations which cause specific triplets (especially those of the form {{font|{{gli|cytosine|C}}n{{gli|guanine|G}}|font=courier|size=big}}) to increase in {{gli|copy number}} above the normal range are highly unstable and responsible for a variety of {{gli|genetic disorders}}.}} |
|||
{{term|triplet}}{{anchor|triplets}} |
|||
{{defn|A unit of three successive {{gli|nucleotides}} in a {{gli|DNA}} or {{gli|RNA}} molecule.<ref name="DoG7"/> A triplet within a coding sequence that codes for a specific amino acid is known as a {{gli|codon}}.}} |
|||
{{term|[[trisomy]]}} |
|||
{{defn|A type of {{gli|polysomy}} in which a {{gli|diploid}} cell or organism has three copies of a particular {{gli|chromosome}} instead of the normal two.}} |
|||
{{term|tRNA}} |
|||
{{defn|See ''{{gli|tRNA|transfer RNA}}''.}} |
|||
{{term|[[true-breeding]]}} |
|||
{{defn|Showing consistent, predictable, and replicable traits in the progeny that result from the mating of any two {{gli|purebred}} parents belonging to the same [[breed]] or variety (and not the traits of other breeds or varieties that may previously have been crossed into the lineage). To "breed true" means that parents of the same lineage will produce offspring that share all of the parents' traits.}} |
|||
{{term|twisting number}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==U== |
|||
{{glossary}} |
|||
{{term|[[ubiquitination]]}} |
|||
{{defn|}} |
|||
{{term|umber}} |
|||
{{defn|See ''{{gli|opal}}''.}} |
|||
{{term|uncharged tRNA}} |
|||
{{defn|A {{gli|tRNA|transfer RNA}} without an attached {{gli|amino acid}}. Contrast ''{{gli|charged tRNA}}''.}} |
|||
{{term|[[underdominance]]}} |
|||
{{defn|}} |
|||
{{term|underwinding}} |
|||
{{defn|See ''{{gli|negative supercoiling}}''.}} |
|||
{{term|[[unequal crossing over]]}} |
|||
{{defn|}} |
|||
{{term|[[uniparental inheritance]]}} |
|||
{{defn|}} |
|||
{{term|unique DNA}} |
|||
{{ghat|Also '''non-repetitive DNA'''.}} |
|||
{{defn|A class of DNA {{gli|nucleic acid sequence|sequences}} determined by {{gli|C0t analysis|C<sub>0</sub>''t'' analysis}} to be present only once in the analyzed genome, as opposed to {{gli|repetitive DNA|repetitive sequences}}. Most structural genes and their introns are unique.<ref name="DoG7"/>}} |
|||
{{term|unstable mutation}} |
|||
{{defn|A {{gli|mutation}} with a high frequency of {{gli|reversion}}.<ref name="DoG7"/>}} |
|||
<span id="untranslated region"></span>{{term|[[untranslated region]] (UTR)}}{{anchor|untranslated regions|UTR|UTRs}} |
|||
{{defn|Either of two non-coding sequences which are transcribed along with a {{gli|protein-coding sequence}}, and thus included within a {{gli|messenger RNA}}, but which are not ultimately {{gli|translated}} during protein synthesis. A typical mRNA transcript includes one such region immediately upstream of the coding sequence, known as the {{gli|5' untranslated region}} (5'-UTR), and one downstream of the coding sequence, known as the {{gli|3' untranslated region}} (3'-UTR). These regions are not removed during {{gli|post-transcriptional modification|post-transcriptional processing}} (unlike {{gli|introns}}) and are usually considered exclusive of the {{gli|5' cap}} and the {{gli|poly(A) tail|3' polyadenylated tail}} (both of which are later additions to a primary transcript and not themselves products of transcription). UTRs are a consequence of the fact that transcription usually begins considerably upstream of the {{gli|start codon}} of the protein-coding sequence and terminates long after the {{gli|stop codon}} has been transcribed, whereas translation is more precise.}} |
|||
{{term|upregulation}} |
|||
{{ghat|Also '''promotion'''.}} |
|||
{{defn|Any process, natural or artificial, which increases the level of {{gli|gene expression}} of a certain {{gli|gene}}. A gene which is observed to be expressed at relatively high levels (such as by detecting higher levels of its {{gli|mRNA}} transcripts) in one sample compared to another sample is said to be ''upregulated''. Contrast ''{{gli|downregulation}}''.}} |
|||
{{term|upstream}} |
|||
{{defn|Towards or closer to the {{gli|5'-end}} of a chain of {{gli|nucleotides}}, or the {{gli|N-terminus}} of a {{gli|peptide}} chain. Contrast ''{{gli|downstream}}''.}} |
|||
<span id="upstream activating sequence"></span>{{term|[[upstream activating sequence]] (UAS)}} |
|||
{{defn|}} |
|||
<span id="uracil"></span>{{term|term=[[uracil]]|content=[[uracil]] ({{font|U|font=courier|size=large}})}} |
|||
{{defn|A {{gli|pyrimidine}} {{gli|nucleobase}} used as one of the four standard nucleobases in {{gli|RNA}} molecules. Uracil forms a {{gli|base pair}} with {{gli|adenine}}. In {{gli|DNA}}, uracil is not used at all, and is instead replaced with {{gli|thymine}}.}} |
|||
<span id="uridine"></span>{{term|term=[[uridine]]|content=[[uridine]] ({{font|U|font=courier|size=large}}, Urd)}} |
|||
{{defn|One of the four standard {{gli|nucleosides}} used in {{gli|RNA}} molecules, consisting of a {{gli|uracil}} {{gli|base}} with its N<sub>9</sub> nitrogen {{gli|glycosidic bond|bonded}} to the C<sub>1</sub> carbon of a {{gli|ribose}} sugar. In {{gli|DNA}}, uridine is replaced with {{gli|thymidine}}.}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==V== |
|||
{{glossary}} |
|||
<span id="VNTR"></span>{{term|[[variable number tandem repeat]] (VNTR)}} |
|||
{{defn|}} |
|||
{{term|[[variegation]]}} |
|||
{{defn|Variation or irregularity in a particular {{gli|phenotype}}, especially a conspicuous visible {{gli|trait}} such as color or pigmentation, occurring simultaneously in different parts of the same individual organism due to any of a variety of causes, such as {{gli|X-inactivation}}, {{gli|mitotic recombination}}, {{gli|transposable element}} activity, {{gli|position-effect variegation|position effects}}, or infection by pathogens.}} |
|||
{{term|[[variome]]}} |
|||
{{defn|}} |
|||
{{term|[[vector (molecular biology)|vector]]}}{{anchor|vectors}} |
|||
{{defn|Any {{gli|DNA}} molecule used as a vehicle to artificially transport foreign genetic material into another cell, where it can be {{gli|DNA replication|replicated}} and/or {{gli|expressed}}. Vectors are typically engineered {{gli|recombinant DNA}} sequences consisting of an {{gli|insertion sequence|insert}} (often a {{gli|transgene}}) and a longer "backbone" sequence containing an {{gli|origin of replication}}, a {{gli|multiple cloning site}}, and a {{gli|selectable marker}}. Vectors are widely used in molecular biology laboratories to isolate, {{gli|molecular cloning|clone}}, or {{gli|expression vector|express}} the insert in the target cell.}} |
|||
{{term|vegetal cell}} |
|||
{{defn|See ''{{gli|somatic cell}}''.}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==W== |
|||
{{glossary}} |
|||
{{term|[[western blotting]]}} |
|||
{{defn|}} |
|||
<span id="WGS"></span>{{term|[[whole genome sequencing]] (WGS)}} |
|||
{{defn|The process of {{gli|sequencing|determining}} the entirety or near-entirety of the DNA sequences comprising an organism's {{gli|genome}} with a single procedure or experiment, generally inclusive of all {{gli|chromosomal}} and {{gli|extrachromosomal DNA|extrachromosomal}} (e.g. {{gli|mtDNA|mitochondrial}}) DNA.}} |
|||
<span id="wild type"></span>{{term|[[wild type]] (WT)}}{{anchor|wild-type}} |
|||
{{ghat|Denoted in shorthand with a <sup>'''''+'''''</sup> superscript.}} |
|||
{{defn|The {{gli|phenotype}} of the typical form of a [[species]] as it occurs in nature; a product of the standard "normal" {{gli|allele}} at a given {{gli|locus}}, as opposed to that produced by a non-standard {{gli|mutant}} allele.}} |
|||
{{term|[[wobble base pair|wobble base pairing]]}} |
|||
{{defn|}} |
|||
{{term|[[writhing number]]}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
{{compact ToC|num=yes|side=yes|top=yes}} |
|||
==X== |
|||
{{glossary}} |
|||
{{term|[[X chromosome]]}} |
|||
{{defn|One of two {{gli|allosome|sex chromosomes}} present in organisms which use the [[XY sex-determination system]], and the only sex chromosome in the [[X0 sex-determination system|X0 system]]. The X chromosome is found in both males and females and typically contains much more {{gli|gene}} content than its counterpart, the {{gli|Y chromosome}}.}} |
|||
{{term|[[X-inactivation]]}} |
|||
{{defn|}} |
|||
{{term|[[X-linked trait]]}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
==Y== |
|||
{{glossary}} |
|||
{{term|[[Y chromosome]]}} |
|||
{{defn|One of two {{gli|allosome|sex chromosomes}} present in organisms which use the [[XY sex-determination system]]. The Y chromosome is found only in males and is typically much smaller than its counterpart, the {{gli|X chromosome}}.}} |
|||
{{term|Y fork}} |
|||
{{defn|See ''{{gli|replication fork}}''.}} |
|||
<span id="YAC"></span>{{term|[[yeast artificial chromosome]] (YAC)}} |
|||
{{defn|}} |
|||
{{glossary end}} |
|||
==Z== |
|||
{{glossary}} |
|||
{{term|[[Z-DNA]]}}{{anchor|Z-DNA}} |
|||
{{defn|}} |
|||
{{term|[[zinc finger]]}} |
|||
{{defn|}} |
|||
{{term|[[zygonema]]}} |
|||
{{ghat|Also '''zygotene stage'''.}} |
|||
{{defn|In {{gli|meiosis}}, the second of five substages of {{gli|prophase|prophase I}}, following {{gli|leptonema}} and preceding {{gli|pachynema}}. During zygonema, {{gli|synapsis}} occurs, physically binding {{gli|homologous chromosomes}} to each other, and the cell's {{gli|centrosome}} divides into two daughter centrosomes, each containing a single {{gli|centriole}}.<ref name="DoG7"/>}} |
|||
{{term|[[zygosity]]}} |
|||
{{defn|The degree to which multiple copies of a {{gli|gene}}, {{gli|chromosome}}, or {{gli|genome}} have the same genetic sequence; e.g. in a diploid organism with two complete copies of its genome (one maternal and one paternal), the degree of similarity of the {{gli|alleles}} present in each copy. Individuals carrying two different alleles for a particular gene are said to be ''{{gli|heterozygous}}'' for that gene; individuals carrying two identical alleles are said to be ''{{gli|homozygous}}'' for that gene. Zygosity may also be considered collectively for a group of genes, or for the entire set of genes and genetic {{gli|loci}} comprising the genome.}} |
|||
{{term|[[zygote]]}} |
|||
{{defn|A type of [[eukaryote|eukaryotic]] cell formed as the direct result of a [[fertilization]] event between two {{gli|gametes}}. In multicellular organisms, the zygote is the earliest developmental stage.}} |
|||
{{glossary end}} |
{{glossary end}} |
||
Revision as of 02:53, 14 November 2022
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology, cell biology, and evolutionary biology.[1] It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.
This glossary is split across two articles:
- Glossary of genetics (0–L) (this page) lists terms beginning with numbers and those beginning with the letters A through L.
- Glossary of genetics (M–Z) lists terms beginning with the letters M through Z.
0–9
- 3' untranslated region (3'-UTR)
- 3'-end
- One of two ends of a single linear strand of DNA or RNA, specifically the end at which the chain of nucleotides terminates at the third carbon atom in the furanose ring of deoxyribose or ribose (i.e. the terminus at which the 3' carbon is not attached to another nucleotide via a phosphodiester bond; in vivo, the 3' carbon is often still bonded to a hydroxyl group). By convention, sequences and structures positioned nearer to the 3'-end relative to others are referred to as downstream. Contrast 5'-end.
- 5' cap
- A specially altered nucleotide attached to the 5'-end of some primary RNA transcripts as part of the set of post-transcriptional modifications which convert raw transcripts into mature RNA products. The precise structure of the 5' cap varies widely by organism; in eukaryotes, the most basic cap consists of a methylated guanine nucleoside bonded to the triphosphate group that terminates the 5'-end of an RNA sequence. Among other functions, capping helps to regulate the export of mature RNAs from the nucleus, prevent their degradation by exonucleases, and promote translation in the cytoplasm. Mature mRNAs can also be decapped.
- 5' untranslated region (5'-UTR)
- 5'-end
- One of two ends of a single linear strand of DNA or RNA, specifically the end at which the chain of nucleotides terminates at the fifth carbon atom in the furanose ring of deoxyribose or ribose (i.e. the terminus at which the 5' carbon is not attached to another nucleotide via a phosphodiester bond; in vivo, the 5' carbon is often still bonded to a phosphate group). By convention, sequences and structures positioned nearer to the 5'-end relative to others are referred to as upstream. Contrast 3'-end.
- 5-bromodeoxyuridine
- See bromodeoxyuridine.
- 5-methyluracil
- See thymine.
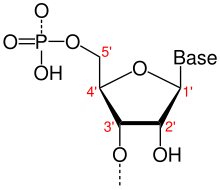
A
- A-DNA
- One of three main biologically active structural conformations of the DNA double helix, along with B-DNA and Z-DNA. The A-form helix has a right-handed twist with 11 base pairs per full turn, only slightly more compact than B-DNA, but its bases are sharply tilted with respect to the helical axis. It is often favored in dehydrated conditions and within sequences of consecutive purine nucleotides (e.g. GAAGGGGA); it is also the primary conformation adopted by double-stranded RNA and RNA-DNA hybrids.[2]
- acentric
- (of a linear chromosome or chromosome fragment) Having no centromere.[3]
- acrocentric
- (of a linear chromosome or chromosome fragment) Having a centromere positioned very close to one end of the chromosome, as opposed to at the end or in the middle.[3]
- activation
- See upregulation.
- activator
- A type of transcription factor that increases the transcription of a gene or set of genes. Most activators work by binding to a specific sequence located within or near an enhancer or promoter and facilitating the binding of RNA polymerase and other transcription machinery in the same region. See also coactivator; contrast repressor.
- adenine (A)
- A purine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Adenine forms a base pair with thymine in DNA and with uracil in RNA.
- adenosine (A)
- One of the four standard nucleosides used in RNA molecules, consisting of an adenine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Adenine bonded to deoxyribose is known as deoxyadenosine, which is the version used in DNA.
- affected relative pair
- Any pair of organisms which are related genetically and both affected by the same trait. For example, two cousins who both have blue eyes are an affected relative pair since they are both affected by the allele that codes for blue eyes.
- allele
- One of multiple alternative versions of an individual gene, each of which is a viable DNA sequence occupying a given position, or locus, on a chromosome. For example, in humans, one allele of the eye-color gene produces blue eyes and another allele of the eye-color gene produces brown eyes.
- allele frequency
- The relative frequency with which a particular allele of a given gene (as opposed to other alleles of the same gene) occurs at a particular locus in the members of a population; more specifically, it is the proportion of all chromosomes within a population that carry a particular allele, expressed as a fraction or percentage. Allele frequency is distinct from genotype frequency, although they are related.
- allosome
- Any chromosome that differs from an ordinary autosome in size, form, or behavior and which is responsible for determining the sex of an organism. In humans, the X chromosome and the Y chromosome are sex chromosomes.
- alpha helix (α-helix)
- A common structural motif in the secondary structures of proteins consisting of a right-handed helix conformation resulting from hydrogen bonding between amino acid residues which are not immediately adjacent to each other.
- alternative splicing
- A regulated phenomenon of eukaryotic gene expression in which specific exons or parts of exons from the same primary transcript are variably included within or removed from the final, mature messenger RNA transcript. A class of post-transcriptional modification, alternative splicing allows a single gene to code for multiple protein isoforms and greatly increases the diversity of proteins that can be produced by an individual genome. See also RNA splicing.
- amber
- One of three stop codons used in the standard genetic code; in RNA, it is specified by the nucleotide triplet UAG. The other two stop codons are named ochre and opal.
- amino acid
- Any of a class of organic compounds whose basic structural formula includes a central carbon atom bonded to amine and carboxyl functional groups and to a variable side chain. Out of nearly 500 known amino acids, a set of 20 are coded for by the standard genetic code and incorporated into long polymeric chains as the building blocks of peptides and hence of polypeptides and proteins. The specific sequences of amino acids in the polypeptide chains that form a protein are ultimately responsible for determining the protein's structure and function.
- amino terminus
- See N-terminus.
- aminoacyl-tRNA (aa-tRNA)
- aminoacyl-tRNA synthetase
- Any of a set of enzymes which catalyze the transesterification reaction that results in the attachment of a specific cognate amino acid (or a precursor) to one of its cognate transfer RNA molecules, forming an aminoacyl-tRNA. Each of the 20 different amino acids used in the genetic code is recognized and attached by its own specific synthetase enzyme, and most synthetases are cognate to several different tRNAs according to their specific anticodons.
- amplicon
- Any DNA or RNA sequence or fragment that is the source and/or product of an amplification reaction. The term is most frequently used to describe the numerous copied fragments that are the products of the polymerase chain reaction or ligase chain reaction, though it may also refer to sequences that are amplified naturally within a genome, e.g. by gene duplication.
- amplification
- The replication of a biomolecule, in particular the production of one or more copies of a nucleic acid sequence, known as an amplicon, either naturally (e.g. by spontaneous duplications) or artificially (e.g. by PCR), and especially implying many repeated replication events resulting in thousands, millions, or billions of copies of the target sequence, which is then said to be amplified.
- anaphase
- The stage of mitosis and meiosis that occurs after metaphase and before telophase, when the replicated chromosomes are segregated and each of the sister chromatids are moved to opposite sides of the cell.
- anaphase lag
- The failure of one or more pairs of sister chromatids or homologous chromosomes to properly migrate to opposite sides of the cell during anaphase of mitosis or meiosis due to a defective spindle apparatus. Consequently, both daughter cells are aneuploid: one is missing one or more chromosomes (creating a monosomy) while the other has one or more extra copies of the same chromosomes (creating a polysomy).
- aneuploidy
- The condition of a cell or organism having an abnormal number of one or more particular chromosomes (but excluding abnormal numbers of complete sets of chromosomes, which instead is known as euploidy).
- annealing
- The hybridization of two single-stranded nucleic acid molecules containing complementary sequences, creating a double-stranded molecule with paired nucleobases. The term is used in particular to describe steps in certain laboratory techniques such as the polymerase chain reaction, where double-stranded DNA molecules are repeatedly denatured into single strands by heating and then exposed to cooler temperatures, causing the strands to reassociate with each other or with complementary primers. The exact temperature at which annealing occurs is strongly influenced by the length and specific sequence of the individual strands.
- anticipation
- A phenomenon by which the symptoms of a genetic disorder become apparent (and often more severe) at an earlier age in affected individuals with each generation that inherits the disorder.
- anticodon
- A series of three consecutive nucleotides within a transfer RNA which complement the three nucleotides of a codon within an mRNA transcript. During translation, each tRNA recruited to the ribosome contains a single anticodon triplet that pairs with one or more complementary codons from the mRNA sequence, allowing each codon to specify a particular amino acid to be added to the growing peptide chain. Anticodons containing inosine in the first position are capable of pairing with more than one codon due to a phenomenon known as wobble base pairing.
- antiparallel
- The orientation of two strands of a double-stranded nucleic acid (and more generally any pair of biopolymers) which are parallel to each other but with opposite directionality. For example, the two complementary strands of a DNA molecule run side-by-side but in opposite directions, with one strand oriented 5'-to-3' and the other 3'-to-5'.
- antisense
- See template strand.
- antisense RNA (asRNA)
- A single-stranded non-coding RNA molecule containing an antisense sequence that is complementary to a sense strand, such as a messenger RNA, with which it readily hybridizes, thereby inhibiting the sense strand's further activity (e.g. translation into protein). Many different classes of naturally occurring RNA such as siRNA function by this principle, making them potent gene silencers in various gene regulation mechanisms. Synthetic antisense RNA has also found widespread use in gene knockdown studies, and in practical applications such as antisense therapy.
- artificial gene synthesis
- A set of laboratory methods used to assemble a gene (or any other nucleic acid sequence) from free nucleotides de novo, i.e. without relying on an existing template strand, which makes it theoretically possible to construct completely synthetic molecules with no restrictions on size or sequence.
- associative overdominance
- The phenomenon by which the linkage of a neutral locus to a selectively maintained polymorphism causes the heterozygosity of the neutral locus to increase.[4]
- asynapsis
- The failure of homologous chromosomes to properly pair with each other during meiosis.[4] Contrast synapsis and desynapsis.
- attached X
- A single monocentric chromosome containing two or more physically attached copies of the normal X chromosome as a result of either a natural internal duplication or any of a variety of genetic engineering methods. The resulting compound chromosome effectively carries two or more doses of all genes and sequences included on the X, yet functions in all other respects as a single chromosome, meaning that haploid 'XX' gametes (rather than the ordinary 'X' gametes) will be produced by meiosis and inherited by progeny. In mechanisms such as genic balance in which the sex of an organism is determined by the total dosage of X-linked genes, an abnormal 'XXY' zygote, fertilized by one XX gamete and one Y gamete, will develop into a female.
- autosome
- Any chromosome that is not an allosome and hence is not involved in the determination of the sex of an organism. Unlike the sex chromosomes, the autosomes in a diploid cell exist in pairs, with the members of each pair having the same structure, morphology, and genetic loci.
- autozygote
- A cell or organism that is homozygous for a locus at which the two homologous alleles are identical by descent, both having been derived from a single gene in a common ancestor.[4] Contrast allozygote.

B
- B-DNA
- The "standard" or classical structural conformation of the DNA double helix in vivo, thought to represent an average of the various distinct conformations assumed by very long DNA molecules under physiological conditions.[2] The B-form double helix has a right-handed twist with a diameter of 23.7 ångströms and a pitch of 35.7 ångströms or about 10.5 base pairs per full turn, such that each nucleotide pair is rotated 36° around the helical axis with respect to its neighboring pairs. See also A-DNA and Z-DNA.
- back mutation
- A mutation that reverses the effect of a previous mutation which had inactivated a gene, thus restoring wild-type function.[5] See also reverse mutation.
- backcrossing
- The breeding of a hybrid organism with one of its parents or an individual genetically similar to one of its parents, often intentionally as a type of selective breeding, with the aim of producing offspring with a genetic identity which is closer to that of the parent. The reproductive event and the resulting progeny are both referred to as a backcross, often abbreviated in genetics shorthand with the symbol BC.
- bacterial artificial chromosome (BAC)
- base
- An abbreviation of nitrogenous base and nucleobase.
- base pair (bp)
- A pair of two nucleobases on complementary DNA or RNA strands which are bonded to each other by hydrogen bonds. The ability of consecutive base pairs to stack one upon another contributes to the long-chain double helix structures observed in both double-stranded DNA and double-stranded RNA molecules.
- baseline
- A measure of the gene expression level of a gene or genes prior to a perturbation in an experiment, as in a negative control. Baseline expression may also refer to the expected or historical measure of expression for a gene.
- bivalent
- blunt end
- bromodeoxyuridine (BUDR, BrdU)

C
- C-terminus
- The end of a linear chain of amino acids (i.e. a peptide) that is terminated by the free carboxyl group (-COOH) of the last amino acid to be added to the chain during translation. This amino acid is said to be C-terminal. By convention, sequences, domains, active sites, or any other structure positioned nearer to the C-terminus of the polypeptide or its folded protein form relative to others are described as downstream. Contrast N-terminus.
- C-value
- The total amount of DNA contained within a haploid nucleus (e.g. a gamete) of a particular organism or species, expressed in number of base pairs or in units of mass (typically picograms); or, equivalently, one-half the amount in a diploid somatic cell. For simple diploid eukaryotes the term is often used interchangeably with genome size, but in certain cases, e.g. in hybrid polyploids descended from parents of different species, the C-value may actually represent two or more distinct genomes contained within the same nucleus. C-values apply only to genomic DNA, and notably exclude extranuclear DNA.
- C-value enigma
- A term used to describe a diverse variety of questions regarding the immense variation in nuclear C-value or genome size among eukaryotic species, in particular the observation that genome size does not correlate with the perceived complexity of organisms, nor necessarily with the number of genes they possess; for example, many single-celled protists have genomes containing thousands of times more DNA than the human genome. This was considered paradoxical until the discovery that eukaryotic genomes consist mostly of non-coding DNA, which lacks genes entirely. The focus of the enigma has since shifted to understanding why and how genomes came to be filled with so much non-coding DNA, and why some genomes have a higher gene content than others.
- cadastral gene
- A regulatory gene that restricts the expression of other genes to specific tissues or body parts in an organism, typically by producing gene products which variably inhibit or permit transcription of the other genes in different cell types.[4] The term is used most commonly in plant genetics.
- canalisation
- The ability of a population to consistently produce the same phenotype regardless of the variability of its environment or the genetic variation within its genome. The concept is most often used in developmental biology to interpret the observation that developmental pathways are frequently shaped by natural selection such that developing cell lineages are "guided" or "canalized" towards a single, definite fate, becoming progressively more resistant to any minor perturbations that may redirect development of the cells away from their initial course.
- candidate gene
- A gene whose location on a chromosome is associated with a particular phenotype (often a disease-related phenotype), and which is therefore suspected of causing or contributing to the phenotype. Candidate genes are often selected for study based on a priori knowledge or speculation about their functional relevance to the trait or disease being researched.
- canonical sequence
- See consensus sequence.
- carboxyl terminus
- See C-terminus.
- carrier
- An individual who has inherited a recessive allele for a genetic trait or mutation but in whom the trait is not usually expressed or observable in the phenotype. Carriers are usually heterozygous for the recessive allele and therefore still able to pass the allele onto their offspring, where the associated phenotype may reappear if the offspring inherits another copy of the allele. The term is commonly used in medical genetics in the context of a disease-causing recessive allele.
- CCAAT box
- A highly conserved regulatory DNA sequence located approximately 75 base pairs upstream (i.e. -75) of the site of the start of transcription for many eukaryotic genes.[3]
- cDNA
- See complementary DNA.
- cell-free DNA (cfDNA)
- Any DNA molecule that exists outside of a cell or nucleus, freely floating in an extracellular fluid such as blood plasma.
- cellular reprogramming
- The conversion of a cell from one tissue-specific cell type to another. This involves dedifferentiation to a pluripotent state; an example is the conversion of mouse somatic cells to an undifferentiated embryonic state, which relies on the transcription factors Oct4, Sox2, Myc, and Klf4.[6]
- centimorgan (cM)
- A unit for measuring genetic linkage defined as the distance between chromosomal loci for which the expected average number of intervening chromosomal crossovers in a single generation is 0.01. Though not an actual measure of physical distance, it is used to infer the actual distance between two loci based on the apparent likelihood of a crossover occurring between them.
- central dogma of molecular biology
- A generalized framework for understanding the flow of genetic information between macromolecules within biological systems. The central dogma outlines the fundamental principle that the sequence information encoded in the three major classes of biopolymer—DNA, RNA, and protein—can only be transferred between these three classes in certain ways, and not in others: specifically, information transfer between the nucleic acids and from nucleic acid to protein is possible, but transfer from protein to protein, or from protein back to either type of nucleic acid, is impossible and does not occur naturally.
- centriole
- A cylindrical organelle composed of microtubules, present only in certain eukaryotes. A pair of centrioles migrate to and define the two opposite poles of a dividing cell where, as part of a centrosome, they initiate the growth of the spindle apparatus.
- centromere
- A specialized DNA sequence within a chromosome that links a pair of sister chromatids. The primary function of the centromere is to act as the site of assembly for kinetochores, protein complexes which direct the attachment of spindle fibers to the centromere and facilitate segregation of the chromatids during mitosis or meiosis.
- centromeric index
- The proportion of the total length of a chromosome encompassed by its short arm, typically expressed as a percentage; e.g. a chromosome with a centromeric index of 15 is acrocentric, with a short arm comprising only 15% of its overall length.[4]
- cfDNA
- See cell-free DNA.
- Chargaff's rules
- A set of axioms which state that, in the DNA of any chromosome, species, or organism, the total number of adenine (A) residues will be approximately equal to the total number of thymine (T) residues, and the number of guanine (G) residues will be equal to the number of cytosine (C) residues; accordingly, the total number of purines (A + G) will equal the total number of pyrimidines (T + C). These observations illustrate the highly specific nature of the complementary base-pairing that occurs in all duplex DNA molecules: even though non-standard pairings are technically possible, they are exceptionally rare because the standard ones are strongly favored in most conditions. Still, the 1:1 equivalence is seldom exact, since at any given time nucleobase ratios are inevitably distorted to some small degree by unrepaired mismatches, missing bases, and non-canonical bases. The presence of single-stranded DNA polymers also alters the proportions, as the sequence of an individual strand may contain any number of any of the bases.
- charged tRNA
- A transfer RNA to which an amino acid has been attached; i.e. an aminoacylated tRNA. Uncharged tRNAs lack amino acids.[4]
- chDNA
- See chloroplast DNA.
- chiasma
- A cross-shaped junction that forms the physical point of contact between two non-sister chromatids belonging to homologous chromosomes during synapsis. As well as ensuring proper segregation of the chromosomes, these junctions are also the breakpoints at which chromosomal crossover may occur during mitosis or meiosis, which results in the reciprocal exchange of DNA between the synapsed chromatids.
- chimerism
- The presence of two or more populations of cells with distinct genotypes in an individual organism, known as a chimera, which has developed from the fusion of cells originating from separate zygotes; each population of cells retains its own genome, such that the organism as a whole is a mixture of genetically non-identical tissues. Genetic chimerism may be inherited (e.g. by the fusion of multiple embryos during pregnancy) or acquired after birth (e.g. by allogeneic transplantation of cells, tissues, or organs from a genetically non-identical donor); in plants, it can result from grafting or errors in cell division. It is similar to but distinct from mosaicism.
- chloroplast DNA (cpDNA, chDNA, ctDNA)
- The set of DNA molecules contained within chloroplasts, a type of photosynthetic plastid organelle located within the cells of some eukaryotes such as plants and algae, representing a semi-autonomous genome separate from that within the cell's nucleus. Like other types of plastid DNA, cpDNA usually exists in the form of small circular plasmids.
- chondriome
- The complete set of mitochondria or of mitochondrial DNA within a cell, tissue, organism, or species.
- chromatid
- One copy of a newly copied chromosome, which is joined to the original chromosome by a centromere.
- chromatin
- A complex of DNA, RNA, and protein found in eukaryotic cells that is the primary substance comprising chromosomes. Chromatin functions as a means of packaging very long DNA molecules into highly organized and densely compacted shapes, which prevents the strands from becoming tangled, reinforces the DNA during cell division, helps to prevent DNA damage, and plays an important role in regulating gene expression and DNA replication.
- chromatin immunoprecipitation (ChIP)
- chromocenter
- A central amorphous mass of polytene chromosomes found in the nuclei of cells of the salivary glands in Drosophila larvae and resulting from the fusion of heterochromatic regions surrounding the centromeres of the somatically paired chromosomes, with the distal euchromatic arms radiating outward.[4]
- chromomere
- A region of a chromosome that has been locally compacted or coiled into chromatin, conspicuous under a microscope as a "bead", node, or dark-staining band, especially when contrasted with nearby uncompacted strings of DNA.
- chromosomal crossover
- chromosomal duplication
- The duplication of an entire chromosome, as opposed to a segment of a chromosome or an individual gene.
- chromosome
- A DNA molecule containing part or all of the genetic material of an organism. Chromosomes may be considered a sort of molecular "package" for carrying DNA within the nucleus of cells and, in most eukaryotes, are composed of long strands of DNA coiled with packaging proteins which bind to and condense the strands to prevent them from becoming an unmanageable tangle. Chromosomes are most easily distinguished and studied in their completely condensed forms, which only occur during cell division. Some simple organisms have only one chromosome made of circular DNA, while most eukaryotes have multiple chromosomes made of linear DNA.
- chromosome condensation
- The process by which eukaryotic chromosomes become shorter, thicker, denser, and more conspicuous under a microscope during prophase due to systemic coiling and supercoiling of chromatic strands of DNA in preparation for cell division.
- chromosome segregation
- The process by which sister chromatids or paired homologous chromosomes separate from each other and migrate to opposite sides of the dividing cell during mitosis or meiosis.
- chromosome walking
- See primer walking.
- circulating tumor DNA (ctDNA)
- Any extracellular DNA fragments derived from tumor cells which are circulating freely in the bloodstream.
- cis
- On the same side; adjacent to; acting from the same molecule. Contrast trans.
- cis-acting
- Affecting a gene or sequence on the same nucleic acid molecule. A locus or sequence within a particular DNA molecule such as a chromosome is said to be cis-acting if it influences or acts upon other sequences located within short distances (i.e. physically nearby, usually but not necessarily downstream) on the same molecule or chromosome; or, in the broadest sense, if it influences or acts upon other sequences located anywhere (not necessarily within a short distance) on the same chromosome of a homologous pair. Cis-acting factors are often involved in the regulation of gene expression by acting to inhibit or to facilitate transcription. Contrast trans-acting.
- cis-dominant mutation
- A mutation occurring within a cis-regulatory element (such as an operator) which alters the functioning of a nearby gene or genes on the same chromosome. Cis-dominant mutations affect the expression of genes because they occur at sites that control transcription rather than within the genes themselves.
- cis-regulatory element (CRE)
- Any sequence or region of non-coding DNA which regulates the transcription of nearby genes (e.g. a promoter, operator, silencer, or enhancer), typically by serving as a binding site for one or more transcription factors. Contrast trans-regulatory.
- cisgenesis
- cistron
- classical genetics
- The branch of genetics based solely on observation of the visible results of reproductive acts, as opposed to that made possible by the modern techniques and methodologies of molecular biology. Contrast molecular genetics.
- cloning
- The process of producing, either naturally or artificially, individual organisms or cells which are genetically identical to each other. Clones are the result of all forms of asexual reproduction, and cells that undergo mitosis produce daughter cells that are clones of the parent cell and of each other. Cloning may also refer to biotechnology methods which artificially create copies of organisms or cells, or, in molecular cloning, copies of DNA fragments or other molecules.
- closed chromatin
- See heterochromatin.
- coactivator
- A type of coregulator that increases the expression of one or more genes by binding to an activator.
- coding strand
- The strand of a double-stranded DNA molecule whose nucleotide sequence corresponds directly to that of the RNA transcript produced during transcription (except that thymine bases are substituted with uracil bases in the RNA molecule). Though it is not itself transcribed, the coding strand is by convention the strand used when displaying a DNA sequence because of the direct analogy between its sequence and the codons of the RNA product. Contrast template strand; see also sense.
- codominance
- codon
- A series of three consecutive nucleotides in a coding region of a nucleic acid sequence. Each of these triplets codes for a particular amino acid or stop signal during protein synthesis. DNA and RNA molecules are each written in a language using four "letters" (four different nucleobases), but the language used to construct proteins includes 20 "letters" (20 different amino acids). Codons provide the key that allows these two languages to be translated into each other. In general, each codon corresponds to a single amino acid (or stop signal). The full set of codons is called the genetic code.
- codon usage bias
- The preferential use of a particular codon to code for a particular amino acid rather than alternative codons that are synonymous for the same amino acid, as evidenced by differences between organisms in the frequencies of the synonymous codons occurring in their coding DNA. Because the genetic code is degenerate, most amino acids can be specified by multiple codons. Nevertheless, certain codons tend to be overrepresented (and others underrepresented) in different species.
- cofactor
- Any non-protein organic compound that is bound to an enzyme. Cofactors are required for the initiation of catalysis.
- comparative genomic hybridization (CGH)
- complementarity
- A property of nucleic acid biopolymers whereby two polymeric chains or "strands" aligned antiparallel to each other will tend to form base pairs consisting of hydrogen bonds between the individual nucleobases comprising each chain, with each type of nucleobase pairing almost exclusively with one other type of nucleobase; e.g. in double-stranded DNA molecules, A pairs only with T and C pairs only with G. Strands that are paired in such a way, and the bases themselves, are said to be complementary. The degree of complementarity between two strands strongly influences the stability of the duplex molecule; certain sequences may also be internally complementary, which can result in a single strand binding to itself. Complementarity is fundamental to the mechanisms governing DNA replication, transcription, and DNA repair.
- complementary DNA (cDNA)
- DNA that is synthesized from a single-stranded RNA template (typically mRNA or miRNA) in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is produced both naturally by retroviruses and artificially in certain laboratory techniques, particularly molecular cloning. In bioinformatics, the term may also be used to refer to the sequence of an mRNA transcript expressed as its DNA coding strand counterpart (i.e. with thymine replacing uracil).
- complementation
- complex trait
- See quantitative trait.
- compound X
- See attached X.
- conditional expression
- The controlled, inducible expression of a transgene, either in vitro or in vivo.
- congression
- The movement of chromosomes to the spindle equator during the prometaphase and metaphase stages of mitosis.[4]
- consanguineous
- (of two or more individuals) Closely genetically related; sharing a recent common ancestor (usually no more than three or four generations distant). The effect of consanguineous mating, also known as inbreeding, is to increase the probability that the progeny will be homozygous at any given pair of genetic loci.[7]
- consensus sequence
- A calculated order of the most frequent residues (of either nucleotides or amino acids) found at each position in a common sequence alignment and obtained by comparing multiple closely related sequence alignments.
- conservation genetics
- An interdisciplinary branch of population genetics which applies genetic methods and concepts in an effort to understand the dynamics of genes in populations, with a principal aim of avoiding extinctions and preserving and restoring biodiversity.
- conservative replication
- conserved sequence
- A nucleic acid or protein sequence that is highly similar or identical across many species or within a genome, indicating that it has remained relatively unchanged through a long period of evolutionary time.
- conspecific
- Belonging to the same species.
- constitutive expression
- 1. The continuous transcription of a gene, as opposed to facultative expression, in which a gene is only transcribed as needed. A gene that is transcribed continuously is called a constitutive gene.
- 2. A gene whose expression depends only on the efficiency of its promoter in binding RNA polymerase,[4] and not on any transcription factors or other regulatory elements which might promote or inhibit its transcription.
- contig
- A continuous sequence of genomic DNA generated by assembling cloned fragments by means of their overlapping sequences.[5]
- copy DNA (cDNA)
- See complementary DNA.
- copy error
- A mutation resulting from a mistake made during DNA replication.[4]
- copy-number variation (CNV)
- A phenomenon in which sections of a genome are repeated and the number of repeats varies between individuals in the population, usually as a result of duplication or deletion events that affect entire genes or sections of chromosomes. Copy-number variations play an important role in generating genetic variation within a population.
- coregulator
- A protein that works together with one or more transcription factors to regulate gene expression.
- corepressor
- A type of coregulator that reduces (represses) the expression of one or more genes by binding to and activating a repressor.
- cosmid
- cpDNA
- See chloroplast DNA.
- CpG site
- A sequence of DNA in which a cytosine nucleotide is immediately followed by a guanine nucleotide on the same strand in the 5'-to-3' direction; the "p" in CpG refers simply to the intervening phosphate group linking the two consecutive nucleotides.
- CpG island
- A region of a genome in which CpG sites occur repetitively or with high frequency.
- crossbreeding
- The breeding of purebred parents belonging to two different breeds, varieties, or populations, often intentionally as a type of selective breeding, with the aim of producing offspring which share traits of both parent lineages or which show heterosis. In animal breeding, the progeny of a cross between breeds of the same species is called a crossbreed, whereas the progeny of a cross between different species is called a hybrid.
- crossing over
- See chromosomal crossover.
- crosslink
- An abnormal chemical bond between two or more nucleobases on opposite strands of a double-stranded DNA molecule (interstrand), or between bases on the same strand (intrastrand), specifically via the formation of covalent bonds that are stronger than the hydrogen bonds of base pairing. Crosslinks can be generated by a variety of exogenous and endogenous agents, and tend to interfere with normal cellular processes such as DNA replication and transcription. They are common targets of DNA repair pathways.
- CRISPR gene editing
- ctDNA
- 1. An abbreviation of circulating tumor DNA.
- 2. An abbreviation of chloroplast DNA.
- cryptic unstable transcript (CUT)
- cytidine (C, Cyd)
- One of the four standard nucleosides used in RNA molecules, consisting of a cytosine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Cytosine bonded to deoxyribose is known as deoxycytidine, which is the version used in DNA.
- cytogenetics
- The branch of genetics that studies how chromosomes influence and relate to cell behavior and function, particularly during mitosis and meiosis.
- cytokinesis
- The final stage of cell division in both mitosis and meiosis, during which the closing of the contractile ring causes the cytoplasm of the parent cell to be divided approximately evenly between the two daughter cells. Contrast karyokinesis.
- cytosine (C)
- A pyrimidine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Cytosine forms a base pair with guanine.

D
- de novo mutation
- A spontaneous mutation in the genome of an individual organism that is new to that organism's lineage, having first appeared in a germ cell of one of the organism's parents or in the fertilized egg that develops into the organism; i.e. a mutation that was not present in either parent's genome.[4]
- degeneracy
- The redundancy of the genetic code, exhibited as the multiplicity of different codons that specify the same amino acid. For example, in the standard genetic code, the amino acid serine is specified by six unique codons (UCA, UCG, UCC, UCU, AGU, and AGC). Codon degeneracy accounts for the existence of synonymous mutations.
- deletion
- A type of mutation in which one or more nucleotides are removed from a nucleic acid sequence.
- denaturation
- The process by which nucleic acids or proteins lose their quaternary, tertiary, and/or secondary structures, either reversibly or irreversibly, through the application of some external chemical or mechanical stress, e.g. by heating, agitation, or exposure to a strong acid or base, all of which can disrupt intermolecular forces such as hydrogen bonding and thereby change or destroy chemical activity. Denatured proteins may be both a cause and a consequence of cell death. Denaturation may also be a normal process; the denaturation of double-stranded DNA molecules, for example, which breaks the hydrogen bonds between base pairs and causes the separation of the duplex molecule into two single strands, is a necessary step in DNA replication and transcription and hence is routinely performed by enzymes such as helicases. The same mechanism is also fundamental to laboratory methods such as PCR.
- deoxyadenosine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of an adenine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Adenine bonded to ribose forms an alternate compound known simply as adenosine, which is used in RNA.
- deoxycytidine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of a cytosine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Cytosine bonded to ribose forms an alternate compound known simply as cytidine, which is used in RNA.
- deoxyguanosine
- One of the four standard deoxyribonucleosides used in DNA molecules, consisting of a guanine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. Guanine bonded to ribose forms an alternate compound known simply as guanosine, which is used in RNA.
- deoxythymidine
- See thymidine.
- deoxyribonuclease (DNase)
- Any of a class of nuclease enzymes which catalyze the hydrolytic cleavage of phosphodiester bonds in DNA molecules, thereby severing strands of deoxyribonucleotides and causing the degradation of DNA polymers into smaller components. Compare ribonuclease.
- deoxyribonucleic acid (DNA)
- A polymeric nucleic acid molecule composed of a series of deoxyribonucleotides, each of which incorporates one of four canonical nucleobases: adenine (A), guanine (G), cytosine (C), and thymine (T). DNA is most often found in double-stranded form, which consists of two complementary antiparallel nucleotide chains in which each of the nucleobases on each individual strand is paired via hydrogen bonding with one on the opposite strand; this structure commonly occurs in the shape of a double helix. DNA can also exist in single-stranded form.
- deoxyribonucleotide
- A nucleotide containing deoxyribose as its pentose sugar component, and the monomeric subunit of deoxyribonucleic acid (DNA) molecules. Deoxyribonucleotides canonically incorporate any of four nitrogenous bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Compare ribonucleotide.
- deoxyribose
- A monosaccharide sugar derived from ribose by the loss of a single oxygen atom. D-deoxyribose, in its pentose ring form, is one of three primary functional groups of deoxyribonucleotides and hence of deoxyribonucleic acid (DNA) molecules.
- desynapsis
- The failure of homologous chromosomes that have synapsed normally during pachynema to remain paired during diplonema. Desynapsis is usually caused by the improper formation of chiasmata.[4] Contrast asynapsis.
- diakinesis
- In meiosis, the fifth and final substage of prophase I, following diplonema and preceding metaphase I. During diakinesis, the chromosomes are further condensed, the two centrosomes reach opposite poles of the cell, and the spindle apparatus begins to extend from the poles to the equator.[4]
- dicentric
- (of a linear chromosome or chromosome fragment) Having two centromeres instead of the normal one.[3]
- dinucleotide
- Any two nucleotides which are immediately adjacent to each other on the same strand of a nucleic acid molecule.
- diploid
- (of a cell or organism) Having two homologous copies of each chromosome. Contrast haploid and polyploid.
- diplonema
- In meiosis, the fourth of the five substages of prophase I, following pachynema and preceding diakinesis. During diplonema, the synaptonemal complex disassembles and the paired homologous chromosomes begin to separate from one another, though they remain tightly bound at the chiasmata where crossover has occurred.
- directionality
- The end-to-end chemical orientation of a single linear strand or sequence of a nucleic acid polymer or a polypeptide. The nomenclature used to indicate nucleic acid directionality is based on the chemical convention of identifying individual carbon atoms in the ribose or deoxyribose sugars of nucleotides, specifically the 5' carbon and 3' carbon of the pentose ring. the sequence of nucleotides in a polymeric chain may be read or interpreted in the 5'-to-3' direction – i.e. starting from the end at which the terminal nucleotide's 5' carbon is not connected to another nucleotide, and proceeding to the end at which the terminal nucleotide's 3' carbon is not connected to another nucleotide – or in the opposite 3'-to-5' direction. Most types of nucleic acid synthesis, including both DNA replication and transcription, work exclusively in the 5'-to-3' direction, because the enzymes involved can only catalyze the addition of free nucleotides to the 3'-end of the previous nucleotide in the chain, and not to the 5'-end. In double-stranded nucleic acids, the two paired strands must be oriented in opposite directions in order to base-pair with each other. Similarly, polypeptide directionality is based on the labeling of the functional groups in amino acids, specifically the amino group, which forms the N-terminus, and the carboxyl group, which forms the C-terminus; amino acid sequences are assembled in the N-to-C direction during translation, and are almost always read or interpreted in the same direction.
- direct repeat
- Any two or more repetitions of a specific sequence of nucleotides occurring in the same orientation (i.e. in precisely the same order and not inverted) and on the same strand, either separated by intervening nucleotides or not. An example is the sequence TACCGnnnnnnTACCG, in which TACCG occurs twice, though separated by six nucleotides that are not part of the repeated sequence. A direct repeat in which the repeats are immediately adjacent to each other is known as a tandem repeat.
- dispersive replication
- distance measure
- Any quantity used to measure the dissimilarity between the gene expression levels of different genes.[8]
- DNA
- See deoxyribonucleic acid.
- DNA barcoding
- A method of taxonomic identification in which short DNA sequences from one or more specific genes are isolated from unidentified samples and then aligned with sequences from a reference library in order to uniquely identify the species or other taxon from which the samples originated. The sequences used in the comparison are chosen carefully from genes that are both widely conserved and that show greater variation between species than within species, e.g. the cytochrome c oxidase gene for eukaryotes or certain ribosomal RNA genes for prokaryotes. These genes are present in nearly all living organisms but tend to evolve different mutations in different species, such that a unique sequence variant can be linked to one particular species, effectively creating a unique identifier akin to a retail barcode. DNA barcoding allows unknown specimens to be identified from otherwise indistinct tissues or body parts, where identification by morphology would be difficult or impossible, and the library of organismal barcodes is now comprehensive enough that even organisms previously unknown to science can often be phylogenetically classified with confidence. The simultaneous identification of multiple different species from a mixed sample is known as metabarcoding.
- DNA condensation
- The process of compacting very long DNA molecules into densely packed, orderly configurations such as chromosomes, either in vivo or in vitro.
- DNA fingerprinting
- DNA microarray
- A high-throughput technology used to measure expression levels of mRNA transcripts or to detect certain changes in nucleotide sequence. It consists of an array of thousands of microscopic spots of DNA oligonucleotides, called features, each containing picomoles of a specific DNA sequence. This can be a short section of a gene or any other DNA element, and is used as a probe to hybridize a cDNA, cRNA or genomic DNA sample (called a target) under high-stringency conditions. Probe-target hybridization is usually detected and quantified by fluorescence-based detection of fluorophore-labeled targets.
- DNA polymerase
- Any of a class of enzymes that synthesizes DNA molecules from individual deoxyribonucleotides. DNA polymerases are essential for DNA replication and usually work in pairs to create identical copies of the two strands of an original double-stranded molecule. They build long chains of DNA by adding nucleotides one at a time to the 3'-end of a DNA strand, usually relying on the template provided by the complementary strand to copy the nucleotide sequence faithfully.
- DNA repair
- The set of processes by which a cell identifies and corrects structural damage or mutations in the DNA molecules that encode its genome. The ability of a cell to repair its DNA is vital to the integrity of the genome and the normal functionality of the organism.
- DNA replication
- The process by which a DNA molecule copies itself, producing two identical copies of one original DNA molecule.
- DNA sequencing
- The process of determining, by any of a variety of different methods and technologies, the order of the bases in the long chain of nucleotides that constitutes a sequence of DNA.
- DNA turnover
- Any mechanism by which DNA sequences are exchanged non-reciprocally (e.g. via gene conversion, transposition, or unequal crossing-over) that causes continual fluctuations in the copy number of DNA motifs during an organism's lifetime. Such mechanisms are often major drivers of speciation between populations.[7]
- DNA-binding domain (DBD)
- DNA-binding protein (DBP)
- Any polypeptide or protein containing one or more domains capable of interacting chemically with one or more parts of a DNA molecule, and consequently having a specific or general affinity for single- and/or double-stranded DNA. DNA-binding activity often depends on the presence and physical accessibility of a specific nucleobase sequence, and mostly occurs at the major groove, since it exposes more of the functional groups that uniquely identify the bases. Binding is also influenced by the spatial conformation of the DNA chain and the occupancy of other proteins near the binding site; many proteins cannot bind to DNA without first undergoing structural changes induced by interactions with other molecules.
- DNase
- See deoxyribonuclease.
- dominance
- A relationship between the alleles of a gene in which one allele produces an effect on phenotype that overpowers or "masks" the contribution of another allele at the same locus; the first allele and its associated phenotypic trait are said to be dominant, and the second allele and its associated trait are said to be recessive. Often, the dominant allele codes for a functional protein while its recessive counterpart does not. Dominance is not an inherent property of any allele or phenotype, but simply describes its relationship to one or more other alleles or phenotypes; it is possible for one allele to be simultaneously dominant over a second allele, recessive to a third, and codominant to a fourth. In genetics shorthand, dominant alleles are often represented by a single uppercase letter (e.g. "A", in contrast to the recessive "a").
- dosage compensation
- Any mechanism by which organisms neutralize the large difference in gene dosage caused by the presence of differing numbers of sex chromosomes in the different sexes, thereby equalizing the expression of sex-linked genes so that the members of each sex receive the same or similar amounts of the products of such genes. An example is X-inactivation in female mammals.
- double helix
- The shape most commonly assumed by double-stranded nucleic acid molecules, resembling a ladder that has been twisted upon its long axis, with the rungs of the ladder consisting of paired nucleobases. This secondary structure is the most energetically stable conformation of the double-stranded forms of both DNA and RNA under most naturally occurring conditions, arising as a consequence of the primary structure of the phosphodiester backbone and the stacking of the nucleotides bonded to it. In B-DNA, the most common DNA variant found in nature, the double helix has a right-handed twist with about 10 base pairs per full turn, and the molecular geometry results in an alternating pattern of "grooves" of differing widths (a major groove and a minor groove) between the parallel backbones.
- double-strand break (DSB)
- The loss of continuity of the phosphate-sugar backbone in both strands of a double-stranded DNA molecule, in particular when the two breaks occur at sites that are directly across from or very close to each other on the complementary strands.[7] Contrast single-strand break.
- double-stranded
- Composed of two antiparallel, complementary nucleic acid molecules or strands (either DNA–DNA, RNA–RNA, or a DNA–RNA hybrid) which are held together by hydrogen bonds between the complementary nucleobases of each strand, known as base pairing. Compare single-stranded.
- double-stranded DNA (dsDNA)
- Any DNA molecule that is composed of two antiparallel, complementary deoxyribonucleotide polymers, known as strands, which are bonded together by hydrogen bonds between the complementary nucleobases. Though it is possible for DNA to exist as a single strand, it is generally more stable and more common in double-stranded form. In most cases, the complementary base pairing causes the twin strands to coil around each other in the shape of a double helix.
- double-stranded RNA (dsRNA)
- Any RNA molecule that is composed of two antiparallel, complementary ribonucleotide polymers, known as strands, which are bonded together by hydrogen bonds between the complementary nucleobases. Though RNA usually occurs in single-stranded form, it is also capable of forming duplexes in the same way as DNA; an example is an mRNA transcript pairing with an antisense version of the same transcript, which effectively silences the gene from which the mRNA was transcribed by preventing translation. As in dsDNA, the base pairing in dsRNA usually causes the twin strands to coil around each other in the shape of a double helix.
- downregulation
- Any process, natural or artificial, which decreases the level of gene expression of a certain gene. A gene which is observed to be expressed at relatively low levels (such as by detecting lower levels of its mRNA transcripts) in one sample compared to another sample is said to be downregulated. Contrast upregulation.
- downstream
- Towards or closer to the 3'-end of a chain of nucleotides, or the C-terminus of a peptide chain. Contrast upstream.
- dsDNA
- See double-stranded DNA.
- dsRNA
- See double-stranded RNA.
- duplex
- See double-stranded.
- duplication
- The production of a second copy of part or all of a nucleotide sequence or amino acid sequence, either naturally or artificially, and the retention of both copies; especially when both the copy and the original sequence are retained in situ within the same molecule, often but not necessarily adjacent to each other. See also gene duplication, chromosomal duplication, and repeat.
- dyad
- See sister chromatids.


E
- ecological genetics
- The study of genetics as it pertains to the ecology and fitness of natural populations of living organisms.
- elongation
- emergenesis
- The quality of genetic traits that results from a specific configuration of interacting genes, rather than simply their combination.
- endonuclease
- Any enzyme whose activity is to cleave phosphodiester bonds within a chain of nucleotides, including those that cleave relatively nonspecifically (without regard to sequence) and those that cleave only at very specific sequences (so-called restriction endonucleases). When recognition of a specific sequence is required, endonucleases make their cuts in the middle of the sequence. Contrast exonuclease.
- enhancer
- A region of DNA near a gene that can be bound by an activator to increase gene expression or by a repressor to decrease expression.
- enhancer RNA (eRNA)
- A subclass of long non-coding RNAs transcribed from regions of DNA containing enhancer sequences. The expression of a given eRNA generally correlates with the activity of the corresponding enhancer in enhancing transcription of its target genes, suggesting that eRNAs play an active role in gene regulation in cis or in trans.
- epigenetics
- epigenome
- episome
- 1. Another name for a plasmid, especially one that is capable of integrating into a chromosome.
- 2. In eukaryotes, any non-integrated extrachromosomal circular DNA molecule that is stably maintained and replicated in the nucleus simultaneously with the rest of the host cell. Such molecules may include viral genomes, bacterial plasmids, and aberrant chromosomal fragments.
- epistasis
- The collective action of multiple genes interacting during gene expression. A form of gene action, epistasis can be either additive or multiplicative in its effects on specific phenotypic traits.
- ergosome
- See polysome.
- euchromatin
- A relatively open, lightly compacted form of chromatin in which DNA is only sporadically bound in nucleosomes and thus broadly accessible to binding and manipulation by proteins and other molecules. Euchromatic regions of a genome are often enriched in genes and actively undergoing transcription, in contrast to heterochromatin, which is relatively gene-poor, nucleosome-rich, and less accessible to transcription machinery.
- euploidy
- The condition of a cell or organism having an abnormal number of complete sets of chromosomes, possibly excluding the sex chromosomes. Euploidy differs from aneuploidy, in which a cell or organism has an abnormal number of one or more specific individual chromosomes.
- evolution
- The change in the heritable characteristics of biological populations over successive generations. In the most traditional sense, it occurs by changes in the frequencies of alleles in a population's gene pool.
- exome
- The entire set of exons within a particular genome, including untranslated regions of mature mRNAs as well as coding regions.
- exon
- Any part of a gene that encodes a part of the final mature messenger RNA produced by that gene after introns have been removed by alternative splicing. The term refers to both the sequence as it exists within a DNA molecule and to the corresponding sequence in RNA transcripts.
- exon skipping
- exonuclease
- Any enzyme whose activity is to cleave phosphodiester bonds within a chain of nucleotides, including those that cleave only upon recognition of a specific sequence (so-called restriction exonucleases). Exonucleases make their cuts at either the 3' or 5'-end of the sequence (rather than in the middle, as with endonucleases).
- exosome complex
- An intracellular multi-protein complex which serves the function of degrading various types of RNA molecules.
- expression vector
- A type of vector, usually a plasmid or viral vector, designed specifically for the expression of a transgene insert in a target cell, rather than for some other purpose such as cloning.
- expressivity
- For a given genotype associated with a variable non-binary phenotype, the proportion of individuals with that genotype who show or express the phenotype to a specified extent, usually given as a percentage. Because of the many complex interactions that govern gene expression, the same allele may produce a wide variety of possible phenotypes of differing qualities or degrees in different individuals; in such cases, both the phenotype and genotype may be said to show variable expressivity. Expressivity attempts to quantify the range of possible levels of phenotypic variation in a population of individuals expressing the phenotype of interest. Compare penetrance.
- extein
- Any part of an amino acid sequence which is retained within a precursor polypeptide, i.e. not excised by post-translational protein splicing, and is therefore present in the mature protein, analogous to the exons of RNA transcripts. Contrast intein.
- extrachromosomal DNA
- Any DNA that is not found in chromosomes or in the nucleus of a cell and hence is not genomic DNA. This may include the DNA contained in plasmids or organelles such as mitochondria or chloroplasts, or, in the broadest sense, DNA introduced by viral infection. Extrachromosomal DNA usually shows significant structural differences from nuclear DNA in the same organism.

F
- facultative expression
- The transcription of a gene only as needed, as opposed to constitutive expression, in which a gene is transcribed continuously. A gene that is transcribed as needed is called a facultative gene.
- fluorescence in situ hybridization (FISH)
- five-prime cap
- See 5' cap.
- five-prime end
- See 5'-end.
- five-prime untranslated region
- See 5' untranslated region.
- fixation
- The process by which a single allele for a particular gene with multiple different alleles increases in frequency in a given population such that it becomes permanently established at 100% frequency – that is, the only allele at that locus within the population's gene pool. In the absence of mutation and heterozygote advantage, any given allele is eventually destined to become either permanently fixed over all other variants or completely lost from the population, though how long this takes depends on selection pressures and chance fluctuations in allele frequencies.
- forward genetics
- forward mutation
- frameshift mutation
- A type of mutation in a nucleic acid sequence caused by the insertion or deletion of a number of nucleotides that is not divisible by three. Because of the triplet nature by which nucleotides code for amino acids, a mutation of this sort causes a shift in the reading frame of the nucleotide sequence, resulting in the sequence of codons downstream of the mutation site being completely different from the original.
- Functional Genomics Data (FGED) Society
- An organization that works with others "to develop standards for biological research data quality, annotation and exchange" as well as software tools that facilitate their use.[9]
G
- G banding
- A technique used in cytogenetics to produce a visible karyotype by staining the condensed chromosomes with Giemsa stain. The staining produces consistent and identifiable patterns of dark and light "bands" in regions of chromatin, which allows specific chromosomes to be easily distinguished.
- gamete
- A haploid cell that is the meiotic product of a progenitor germ cell and the final product of the germ line in sexually reproducing multicellular organisms. Gametes are the means by which an organism passes its genetic information to its offspring; during fertilization, two gametes (one from each parent) are fused into a single diploid zygote.
- gDNA
- See genomic DNA.
- gene
- Any segment or set of segments of a nucleic acid molecule that contains the information necessary to produce a functional RNA transcript in a controlled manner. In living organisms, genes are often considered the fundamental units of heredity and are typically encoded in DNA. A particular gene can have multiple different versions, or alleles, and a single gene can result in a gene product that influences many different phenotypes.
- gene dosage
- The number of copies of a particular gene present in a genome. Gene dosage directly influences the amount of gene product a cell is able to express, though a variety of controls have evolved which tightly regulate gene expression. Changes in gene dosage caused by mutations include copy-number variations.
- gene drive
- gene duplication
- A type of mutation defined as any duplication of a region of DNA that contains a gene. Compare chromosomal duplication.
- gene expression
- The set of processes by which the information encoded in a gene is used in the synthesis of a gene product, such as a protein or non-coding RNA, or otherwise made available to influence one or more phenotypes. Canonically, the first step is transcription, which produces a messenger RNA molecule complementary to the DNA molecule in which the gene is encoded; for protein-coding genes, the second step is translation, in which the messenger RNA is read by the ribosome to produce a protein. The information contained within a DNA sequence need not necessarily be transcribed and translated to exert an influence on molecular events, however; broader definitions encompass a huge variety of other ways in which genetic information can be expressed.
- Gene Expression Omnibus (GEO)
- A database of high-throughput functional genomics and gene expression data derived from experimental chips and next-generation sequencing and managed by the National Center for Biotechnology Information.[10][11]
- gene fusion
- The union, either by natural mutation or by recombinant laboratory techniques, of two or more previously independent genes that code for different gene products such that they become subject to control by the same regulatory systems. The resulting hybrid sequence is known as a fusion gene.[4]
- gene mapping
- Any of a variety of methods used to precisely identify the location of a particular gene within a DNA molecule (such as a chromosome) and/or the physical or linkage distances between it and other genes.
- gene of interest (GOI)
- gene pool
- The sum of all of the various alleles shared by the members of a single population.
- gene product
- Any of the biochemical material resulting from the expression of a gene, most commonly interpreted as the functional mRNA transcript produced by transcription of the gene or the fully constructed protein produced by translation of the transcript, though non-coding RNA molecules such as transfer RNAs may also be considered gene products. A measurement of the quantity of a given gene product that is detectable in a cell or tissue is sometimes used to infer how active the corresponding gene is.
- gene regulation
- The broad range of mechanisms used by cells to control the activity of their genes, especially to allow, prohibit, increase, or decrease the production or expression of specific gene products, such as RNA or proteins. Gene regulation increases an organism's versatility and adaptability by allowing its cells to express different gene products when required by changes in its environment. In multicellular organisms, the regulation of gene expression also drives cellular differentiation and morphogenesis in the embryo, enabling the creation of a diverse array of cell types from the same genome.
- gene silencing
- Any mechanism of gene regulation which drastically reduces or completely prevents the expression of a particular gene. Gene silencing may occur naturally during either transcription or translation. Laboratory techniques often exploit natural silencing mechanisms to achieve gene knockdown.
- gene therapy
- The insertion of a functional or wild-type gene or part of a gene into an organism (especially a patient) with the intention of correcting a genetic defect, either by direct substitution of the defective gene or by supplementation with a second, functional version.[7]
- gene trapping
- A high-throughput technology used to simultaneously inactivate, identify, and report the expression of a target gene in a mammalian genome by introducing an insertional mutation consisting of a promoterless reporter gene and/or a selectable genetic marker flanked by an upstream splice site and a downstream polyadenylated termination sequence.
- generation
- 1. In any given organism, a single reproductive cycle, or the phase between two consecutive reproductive events, i.e. between an individual organism's reproduction and that of the progeny of that reproduction; or the actual or average length of time required to complete a single reproductive cycle, either for a particular lineage or for a population or species as a whole.
- 2. In a given population, those individuals (often but not necessarily living contemporaneously) who are equally removed from a given common ancestor by virtue of the same number of reproductive events having occurred between them and the ancestor.[7]
- genetic association
- The co-occurrence within a population of one or more alleles or genotypes with a particular phenotypic trait more often than might be expected by chance alone; such statistical correlation may be used to infer that the alleles or genotypes are responsible for producing the given phenotype.
- genetic background
- genetic code
- A set of rules by which information encoded within nucleic acids is translated into proteins by living cells. These rules define how sequences of nucleotide triplets called codons specify which amino acid will be added next during protein synthesis. The vast majority of living organisms use the same genetic code (sometimes referred to as the "standard" genetic code) but variant codes do exist.
- genetic counseling
- The process of advising individuals or families who are affected by or at risk of developing genetic disorders in order to help them understand and adapt to the physiological, psychological, and familial implications of genetic contributions to disease. Genetic counseling integrates genetic testing, genetic genealogy, and genetic epidemiology.[12]
- genetic disorder
- genetic distance
- A measure of the genetic divergence between species, populations within a species, or individuals, used especially in phylogenetics to express either the time elapsed since the existence of a common ancestor or the degree of differentiation in the DNA sequences comprising the genomes of each population or individual.
- genetic diversity
- The total number of genetic traits or characteristics in the genetic make-up of a population, species, or other group of organisms. It is often used as a measure of the adaptability of a group to changing environments. Genetic diversity is similar to, though distinct from, genetic variability.
- genetic drift
- A change in the frequency with which an existing allele occurs in a population due to random variation in the distribution of alleles from one generation to the next. Genetic drift is often interpreted as the role that random chance plays in determining whether a given allele becomes more or less common with each generation, regardless of the influence of natural selection. Such randomness may cause certain alleles, even otherwise advantageous ones, to disappear completely from the gene pool, thereby reducing genetic variation, or it may cause initially rare alleles, even neutral or deleterious ones, to become much more frequent or even fixed.
- genetic engineering
- The direct, deliberate manipulation of an organism's genetic material using any of a variety of biotechnology methods, including the insertion or removal of genes, the transfer of genes within and between species, the mutation of existing sequences, and the construction of novel sequences using artificial gene synthesis. Genetic engineering encompasses a broad set of technologies by which the genetic composition of individual cells, tissues, or entire organisms may be altered for various purposes, commonly in order to study the functions and expression of individual genes, to produce hormones, vaccines, and other drugs, and to create genetically modified organisms for use in research and agriculture.
- genetic epidemiology
- The study of the role played by genetic factors in determining health and disease, in particular through the interaction of genetic factors with environmental factors, and typically as observed in genetically related individuals, often families or lineages but also populations and subpopulations.
- genetic genealogy
- The use of genealogical DNA testing in combination with traditional genealogical methods to infer the level and type of genetic relationships between individuals, to find ancestors, and to construct family trees, genograms, or other genealogical charts.
- genetic hitchhiking
- A type of linked selection by which the positive selection of an allele undergoing a selective sweep causes alleles for different genes at nearby loci to change frequency as well, allowing them to "hitchhike" to fixation along with the positively selected allele. If selection at the first locus is strong enough, neutral or even slightly deleterious alleles within the same linkage group may undergo the same positive selection because the physical distance between the nearby loci is small enough that a recombination event is unlikely to occur between them. Genetic hitchhiking is often considered the opposite of background selection.
- genetic marker
- A specific, easily identifiable, and usually highly polymorphic gene or other DNA sequence with a known location on a chromosome that can be used to identify the individual or species possessing it.
- genetic recombination
- Any reassortment or exchange of genetic material within an individual organism or between individuals of the same or different species, especially that which creates genetic variation. In the broadest sense, the term encompasses a diverse class of naturally occurring mechanisms by which nucleic acid sequences are copied or physically transferred into different genetic environments, including homologous recombination during meiosis or mitosis or as a normal part of DNA repair; horizontal gene transfer events such as bacterial conjugation, viral transduction, or transformation; or errors in DNA replication or cell division. Artificial recombination is central to many genetic engineering techniques which produce recombinant DNA.
- genetic redundancy
- The redundant encoding of two or more distinct gene products that ultimately perform the same biochemical function. Mutations in one of these genes may have a smaller effect on fitness than might be expected, since the redundant genes often compensate for any loss of function and obviate any gain of function.
- genetic regulatory network (GRN)
- A graph that represents the regulatory complexity of gene expression. The vertices (nodes) are represented by various regulatory elements and gene products while the edges (links) are represented by their interactions. These network structures also represent functional relationships by approximating the rate at which genes are transcribed.
- genetic testing
- A broad class of various procedures used to identify features of an individual's particular chromosomes, genes, or proteins in order to determine parentage or ancestry, diagnose vulnerabilities to heritable diseases, or detect mutant alleles associated with increased risks of developing genetic disorders. Genetic testing is widely used in human medicine, agriculture, and biological research.
- genetic variability
- The formation or the presence of individuals differing in genotype within a population or other group of organisms, as opposed to individuals with environmentally induced differences, which cause only temporary, non-heritable changes in phenotype. Barring other limitations, a population with high genetic variability has a greater potential for successful adaptation to changing environmental conditions than a population with low genetic variability. Genetic variability is similar to, though distinct from, genetic diversity.
- genetic variation
- The genetic differences both within and between populations, species, or other groups of organisms. It is often visualized as the variety of different alleles in the gene pools of different populations.
- genetically modified organism (GMO)
- Any organism whose genetic material has been altered using genetic engineering techniques, particularly in a way that does not occur naturally by mating or by natural genetic recombination.
- genetics
- The field of biology that studies genes, genetic variation, and heredity in living organisms.
- genic balance
- A mechanism of sex determination that depends upon the ratio of the number of X chromosomes (X) to the number of sets of autosomes (A). Males develop when the X/A ratio is 0.5 or less, females when it is 1.0 or more, and an intersex develops when it is between 0.5 and 1.0.[4]
- genome
- The entire complement of genetic material contained within the chromosomes of an organism, organelle, or virus. The term is also used to refer to the collective set of genetic loci shared by every member of a population or species, regardless of the different alleles that may be present at these loci in different individuals.
- genome size
- The total amount of DNA contained within one copy of a genome, typically measured by mass (in picograms or daltons) or by the total number of base pairs (in kilobases or megabases). For diploid organisms, genome size is often used interchangeably with C-value.
- genomic DNA (gDNA)
- The DNA contained in chromosomes, as opposed to the extrachromosomal DNA contained in separate structures such as plasmids or organelles such as mitochondria or chloroplasts.
- genomic imprinting
- An epigenetic phenomenon that causes genes to be expressed in a manner dependent upon the particular parent from which the gene was inherited. It occurs when epigenetic marks such as DNA or histone methylation are established or "imprinted" in the germ cells of a parent organism and subsequently maintained through cell divisions in the somatic cells of the organism's progeny; as a result, a gene in the progeny that was inherited from the father may be expressed differently than another copy of the same gene that was inherited from the mother.
- genomics
- An interdisciplinary field that studies the structure, function, evolution, mapping, and editing of entire genomes, as opposed to individual genes.
- genotoxicity
- The ability of certain chemical agents to cause damage to genetic material within a living cell (e.g. through single- or double-stranded breaks, crosslinking, or point mutations), which may or may not result in a permanent mutation. Though all mutagens are genotoxic, not all genotoxic compounds are mutagenic.
- genotype
- The entire complement of alleles present in a particular individual's genome, which gives rise to the individual's phenotype.
- genotype frequency
- genotyping
- The process of determining differences in the genotype of an individual by examining the DNA sequences in the individual's genome using bioassays and comparing them to another individual's sequences or a reference sequence.
- germ cell
- Any cell that gives rise to the gametes of an organism that reproduces sexually. Germ cells are the vessels for the genetic material which will ultimately be passed on to the organism's descendants and are usually distinguished from somatic cells, which are entirely separate from the germ line.
- germ line
- 1. In multicellular organisms, the population of cells which are capable of passing on their genetic material to the organism's progeny and are therefore (at least theoretically) distinct from somatic cells, which cannot pass on their genetic material except to their own immediate mitotic daughter cells. Cells of the germ line are called germ cells.
- 2. The lineage of germ cells, spanning many generations, that contains the genetic material which has been passed on to an individual from its ancestors.
- Goldberg-Hogness box
- See TATA box.
- gRNA
- See guide RNA.
- guanine (G)
- A purine nucleobase used as one of the four standard nucleobases in both DNA and RNA molecules. Guanine forms a base pair with cytosine.
- guanine-cytosine content
- The proportion of nitrogenous bases in a nucleic acid that are either guanine (G) or cytosine (C), typically expressed as a percentage. DNA and RNA molecules with higher GC-content are generally more thermostable than those with lower GC-content due to molecular interactions that occur during base stacking.[13]
- guanosine (G, Guo)
- One of the four standard nucleosides used in RNA molecules, consisting of a guanine base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. Guanine bonded to deoxyribose is known as deoxyguanosine, which is the version used in DNA.
- guide RNA (gRNA)
H
- hairpin
- See stem-loop.
- haplodiploidy
- A sex-determination system in which the sex of an individual organism is determined by the number of sets of chromosomes it possesses: offspring which develop from fertilized eggs are diploid and female, while offspring which develop from unfertilized eggs are haploid and male, with half as many chromosomes as the females. Haplodiploidy is common to all members of the insect order Hymenoptera and several other insect taxa.
- haplogroup
- haploid
- (of a cell or organism) Having one copy of each chromosome, with each copy not being part of a pair. Contrast diploid and polyploid.
- haploinsufficiency
- haplotype
- A set of alleles in an individual organism that were inherited together from a single parent.
- helicase
- Any of a class of ATP-dependent motor proteins that move directionally along the DNA backbone and catalyze the separation of the two complementary strands of double-stranded molecules, permitting a wide variety of vital processes to take place, e.g. transcription, replication, and repair.[7]
- hemizygous
- In a diploid organism, having just one allele at a given genetic locus (where there would ordinarily be two). Hemizygosity may be observed when only one copy of a chromosome is present in a normally diploid cell or organism, or when a segment of a chromosome containing one copy of an allele is deleted, or when a gene is located on a sex chromosome in the heterogametic sex (in which the sex chromosomes do not exist in matching pairs); for example, in human males with normal chromosomes, almost all X-linked genes are said to be hemizygous because there is only one X chromosome and few of the same genes exist on the Y chromosome.
- heredity
- The storage, transfer, and expression of molecular information in biological organisms,[7] as manifested by the passing on of phenotypic traits from parents to their offspring, either through sexual or asexual reproduction. Offspring cells or organisms are said to inherit the genetic information of their parents.
- heritability
- 1. The ability to be inherited.
- 2. A statistic used in quantitative genetics that estimates the proportion of variation within a given phenotypic trait that is due to genetic variation between individuals in a particular population. Heritability is estimated by comparing the individual phenotypes of closely related individuals in the population.
- heterochromatin
- heterochromosome
- See allosome.
- heterogeneous nuclear RNA (hnRNA)
- heterologous expression
- The expression of a foreign gene or any other foreign DNA sequence within a host organism which does not naturally contain the same gene. Insertion of foreign transgenes into heterologous hosts using recombinant vectors is a common biotechnology method for studying gene structure and function.
- heterosis
- The improved or increased function or quality of any biological trait in a hybrid offspring, with respect to the same trait in its genetically distinct parents. If any one or more of the parents' traits are noticeably enhanced in the offspring as a result of the mixing of the parents' genetic contributions, the offspring is said to be heterotic.
- heterozygous
- In a diploid organism, having two different alleles at a given genetic locus. In genetics shorthand, heterozygous genotypes are represented by a pair of non-matching letters or symbols, often an uppercase letter (indicating a dominant allele) and a lowercase letter (indicating a recessive allele), such as "Aa" or "Bb". Contrast homozygous.
- high-throughput
- histone
- Any of a class of highly alkaline proteins responsible for packaging nuclear DNA into structural units called nucleosomes in eukaryotic cells. Histones are the chief protein components of chromatin, where they associate into complexes which act as "spools" around which the linear DNA molecule winds. They play a major role in gene regulation and expression.
- hnRNA
- See heterogeneous nuclear RNA.
- holocentric
- (of a linear chromosome or chromosome fragment) Having no single centromere but rather multiple kinetochore assembly sites dispersed along the entire length of the chromosome. During cell division, the chromatids of holocentric chromosomes move apart in parallel and do not form the classical V-shaped structures typical of monocentric chromosomes.
- homeobox
- homologous chromosomes
- A set of two matching chromosomes, one maternal and one paternal, which pair up with each other inside the nucleus during meiosis. They have the same genes at the same loci, but may have different alleles.
- homologous recombination
- A type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical ("homologous") molecules of DNA, especially that which occurs between homologous chromosomes. The term may refer to the recombination that occurs as a part of any of a number of distinct cellular processes, most commonly DNA repair or chromosomal crossover during meiosis in eukaryotes and horizontal gene transfer in prokaryotes. Contrast nonhomologous recombination.
- homozygous
- In a diploid organism, having two identical alleles at a given genetic locus. In genetics shorthand, homozygous genotypes are represented by a pair of matching letters or symbols, such as "AA" or "aa". Contrast heterozygous.
- horizontal gene transfer (HGT)
- Any process by which genetic material is transferred between unicellular and/or multicellular organisms other than by vertical transmission from parent to offspring.
- housekeeping gene
- Any constitutive gene that is transcribed at a relatively constant level across many or all known conditions. Such a gene's products typically play critical roles in the maintenance of basic cellular function. It is generally assumed that their expression is unaffected by experimental or pathological conditions.
- Hox genes
- Human Genome Project (HGP)
- hybrid
- The offspring that results from combining the qualities of two organisms of different genera, species, breeds, or varieties through sexual reproduction. Hybrids may occur naturally or artificially, as during selective breeding of domesticated animals and plants. Reproductive barriers typically prevent hybridization between distantly related organisms, or at least ensure that hybrid offspring are sterile, but fertile hybrids may result in speciation.
- hybridization
- 1. The process by which a hybrid organism is produced from two organisms of different genera, species, breeds, or varieties.
- 2. The process by which two or more single-stranded nucleic acid molecules with complementary nucleotide sequences pair with each other in solution, creating double-stranded or triple-stranded molecules via the formation of hydrogen bonds between the complementary nucleobases of each strand. In certain laboratory contexts, especially ones in which long strands hybridize with short oligonucleotide primers, hybridization is often referred to as annealing.
- 3. A step in some experimental assays in which a single-stranded DNA or RNA preparation is added to an array surface and anneals to a complementary probe.
- hybridization probe
- hypoxanthine (I)
- A naturally occurring non-canonical purine nucleobase that is used in some RNA molecules and pairs with standard nucleobases in a phenomenon known as wobble base pairing. Its nucleoside form is known as inosine, which is the reason it is commonly abbreviated with the letter I in sequence reads.
I
- idiochromosome
- See allosome.
- idiomere
- See chromomere.
- in silico
- (of a scientific experiment or research) Conducted, produced, or analyzed by means of computer modeling or simulation, as opposed to a real-world trial.
- in situ
- (of a scientific experiment or biological process) Occurring or made to occur in a natural, uncontrolled setting, or in the natural or original position or place, as opposed to in a foreign cell or tissue type or in an artificial environment.
- in situ hybridization
- in vitro
- (of a scientific experiment or biological process) Occurring or made to occur in a laboratory vessel or other controlled artificial environment, e.g. in a test tube or a petri dish, as opposed to inside a living organism or in a natural setting.
- in vivo
- (of a scientific experiment or biological process) Occurring or made to occur inside the cells or tissues of a living organism; or, in the broadest sense, in any natural, unmanipulated setting. Contrast ex vivo and in vitro.
- inbred line
- Any lineage of a particular species in which individuals are nearly or completely genetically identical to each other due to a long history of repeated inbreeding, either by natural or artificial means. Lineages are typically considered inbred after at least 20 generations of inbreeding (e.g. by self-fertilization or sib mating), at which point nearly all loci across the genome are homozygous and all individuals can therefore effectively be treated as clones (despite the fact that individuals are still produced by sexual reproduction).
- inbreeding
- Sexual reproduction between breeds or individuals that are closely related genetically. Inbreeding results in homozygosity, which can increase both the probability of offspring being affected by deleterious recessive traits and the probability of fixing beneficial traits within the breeding population. The reproductive event and the resulting progeny may both be referred to as an incross, and the progeny is said to be inbred. Contrast outbreeding.
- incidence
- The frequency of new occurrence of a genetic disorder (or more broadly any genetic condition or trait, deleterious or otherwise) among the members of a particular population and within a particular period of time.[7]
- incomplete dominance
- indel
- A term referring to either an insertion or a deletion of one or more bases in a nucleic acid sequence.
- inducer
- A protein that binds to a repressor (to disable it) or to an activator (to enable it).
- inducible gene
- A gene whose expression is either responsive to environmental change or dependent on its host cell's position within the cell cycle.
- inheritance
- See heredity.
- initiation codon
- See start codon.
- inosine
- insertion
- A type of mutation in which one or more bases are added to a nucleic acid sequence. Contrast deletion.
- insertion sequence (IS)
- Any nucleotide sequence that is inserted naturally or artificially into another sequence. The term is used in particular to refer to the part of a transposable element that codes for those proteins directly involved in the transposition process, e.g. the transposase enzyme. The coding region in a transposable insertion sequence is usually flanked by short inverted repeats, and the structure of larger transposable elements may include a pair of flanking insertion sequences which are themselves inverted.
- insertional mutagenesis
- The alteration of a DNA sequence by the insertion of one or more nucleotides into the sequence, either naturally or artificially. Depending on the precise location of the insertion within the target sequence, insertions may partially or totally inactivate or even upregulate a gene product or biochemical pathway, or they may be neutral, leading to no substantive changes at all. Many genetic engineering techniques rely on the insertion of exogenous genetic material into host cells in order to study gene function and expression.[4]
- insulator
- A specific DNA sequence that prevents a gene from being influenced by the activation or repression of nearby genes.
- integron
- A mobile genetic element consisting of a gene cassette that encodes a site-specific recombinase and includes integrase-specific recognition sites and a promoter that governs the expression of one or more genes conferring adaptive traits on the host cell. Integrons usually exist in the form of circular episomal DNA fragments such as plasmids, through which they aid the rapid adaptation and evolution of bacteria by enabling horizontal gene transfer of antibiotic resistance genes between different bacterial species.[4]
- intein
- Any sequence of one or more amino acids within a precursor polypeptide that is excised by protein splicing during post-translational modification and is therefore absent from the mature protein, analogous to the introns spliced out of RNA transcripts.[4] Contrast extein.
- intercalating agent
- Any chemical compound (e.g. acridine dyes) that disrupts the alignment and pairing of bases in the complementary strands of a DNA molecule by inserting itself between the bases.[3]
- intercalation
- The insertion, naturally or artificially, of chemical compounds between the planar bases of a DNA molecule, which generally disrupts the hydrogen bonding necessary for base pairing.
- intercistronic region
- Any DNA sequence that is located between the stop codon of one gene and the start codon of the following gene in a polycistronic transcription unit.[4] See also intergenic region.
- intercross
- A cross in which both the male and female parents are heterozygous at a particular locus.[4]
- intergenic region (IGR)
- Any sequence of non-coding DNA that is located between functional genes.
- intergenic spacer (IGS)
- See spacer.
- interkinesis
- The abbreviated pause in activities related to cell division that occurs during meiosis in some species, between the first and second meiotic divisions (i.e. meiosis I and meiosis II). No DNA replication occurs during interkinesis, unlike during the normal interphase that precedes meiosis I and mitosis.[4]
- interphase
- All stages of the cell cycle excluding cell division. A typical cell spends most of its life in interphase, during which it conducts everyday metabolic activities as well as the complete replication of its genome in preparation for mitosis or meiosis.
- intragenic region
- See intron.
- intragenic suppression
- intragenomic conflict
- introgression
- The movement of a gene from the gene pool of one population or species into that of another population by the repeated backcrossing of hybrids of the two populations with one of the parent populations. Introgression is a ubiquitous and important source of genetic variation in natural populations, but may also be practiced intentionally in the cultivation of domesticated plants and animals.
- intron
- Any nucleotide sequence within a functional gene that is removed by RNA splicing during post-transcriptional modification of the mRNA primary transcript and is therefore absent from the final mature mRNA. The term refers to both the sequence as it exists within a DNA molecule and to the corresponding sequence in RNA transcripts. Contrast exon.
- intron intrusion
- intron-mediated recombination
- See exon shuffling.
- inverted repeat
- A nucleotide sequence followed downstream on the same strand by its own reverse complement. The initial sequence and the reverse complement may be separated by any number of nucleotides, or may be immediately adjacent to each other; in the latter case, the composite sequence is also called a palindromic sequence. Inverted repeats are self-complementary by definition, a property which involves them in many important biological functions and dysfunctions. Contrast direct repeat.
- in-frame
- 1. (of a gene or sequence) Read or transcribed in the same reading frame as another gene or sequence; not requiring a shift in reading frame to be intelligible or to result in a functional peptide.
- 2. (of a mutation) Not causing a frameshift.[4]
- isochore
- isochromosome
- A type of abnormal chromosome in which the arms of the chromosome are mirror images of each other. Isochromosome formation is equivalent to simultaneous duplication and deletion events such that two copies of either the long arm or the short arm comprise the resulting chromosome.
- isomeric genes
- Two or more genes that are equivalent and redundant in the sense that, despite coding for distinct gene products, they each result in the same phenotype when set within the same genetic background. If several isomeric genes are present in a single genotype they may be either cumulative or non-cumulative in their contributions to the phenotype.[7]
J
- jumping gene
- See transposable element.
- junctional diversity
- junk DNA
- Any DNA sequence that appears to have no known biological function, or which serves a purpose that has no positive or a net negative effect on the fitness of the genome in which it is located. The term was once more broadly used to refer to all non-coding DNA, much of which was later discovered to have a function; in modern usage it typically refers to broken or vestigial sequences and selfish genetic elements, including introns, pseudogenes, intergenic DNA, and fragments of transposons and retroviruses, which together constitute a large proportion of the genomes of most eukaryotes. Despite not contributing productively to the host organism, these sequences are able to persist indefinitely inside genomes because the disadvantages of continuing to copy them are too small to be acted upon by natural selection.
- junk RNA
- Any RNA-encoded sequence, especially a transcript, that appears to have no known biological function, or whose function has no positive or a net negative effect on the fitness of the genome from which it is transcribed. Despite remaining untranslated, many non-coding RNAs still serve important functions, whereas junk RNAs are truly useless: often they are the product of accidental transcription of a junk DNA sequence, or they may result from post-transcriptional processing of primary transcripts, as with spliced-out introns. Junk RNA is usually quickly degraded by ribonucleases and other cytoplasmic enzymes.
K
- Ka/Ks ratio
- karyotype
- The number and appearance of chromosomes within the nucleus of a eukaryotic cell, especially as depicted in an organized photomicrograph known as a karyogram or idiogram (in pairs and ordered by size and by position of the centromere). The term is also used to refer to the complete set of chromosomes in a species or individual organism or to any test that detects this complement or measures the chromosome number.
- kilobase (kb)
- A unit of nucleic acid length equal to 1,000 base pairs in duplex molecules such as double-stranded DNA or 1,000 bases in single-stranded molecules.
- kinetochore
- A disc-shaped protein complex which assembles around the centromere of a chromosome during prometaphase of mitosis and meiosis, where it functions as the attachment point for microtubules of the spindle apparatus.
- knob
- In cytogenetics, an enlarged, heavily staining chromomere that can be used as a visual marker, allowing specific chromosomes to be easily identified in the nucleus.[4]
- knockdown
- A genetic engineering technique by which the normal rate of expression of one or more of an organism's genes is reduced, either through direct modification of a DNA sequence or through treatment with a reagent such as a short DNA or RNA oligonucleotide with a sequence complementary to either an mRNA transcript or a gene.
- knockin
- knockout
- A genetic engineering technique in which an organism is modified to carry genes that have been made inoperative ("knocked out"), such that their expression is disrupted at some point in the pathway that produces their gene products and the organism is deprived of their normal effects. Contrast knockin.

L
- labelling
- A laboratory technique involving the chemical attachment of a highly selective substance, known as a label, tag, or probe, to a particular cell, protein, amino acid, or other biomolecule of interest, either in vivo or in vitro. The label is typically a reactive derivative of a naturally fluorescent compound (e.g. green fluorescent protein) or any other substance that makes its target distinguishable in some way; other commonly used labels include dyes, enzymes, antibodies, and radioactive molecules. The labelled targets are thereby rendered distinct from their surroundings, allowing them to be easily detected, identified, quantified, or isolated for further study.
- lagging strand
- In DNA replication, the nascent strand for which DNA polymerase's direction of synthesis is away from the replication fork, which necessitates a complex and discontinuous process, in contrast to the streamlined, continuous synthesis of the other nascent strand that occurs simultaneously. Because DNA polymerase works only in the 5' to 3' direction, but the overall direction of chain elongation must ultimately be the opposite (i.e. 3' to 5', toward the replication fork), the elongation must occur by an indirect mechanism in which a primase synthesizes short RNA primers complementary to the template DNA, and DNA polymerase then extends the primed segments into short chains of nucleotides known as Okazaki fragments. The RNA primers are then removed and replaced with DNA, and the Okazaki fragments are joined by DNA ligase.
- lampbrush chromosome
- lateral gene transfer (LGT)
- See horizontal gene transfer.
- Law of Dominance and Uniformity
- One of three fundamental principles of Mendelian inheritance, which states that different alleles of the same gene may be dominant or recessive relative to others, and that an organism with at least one dominant allele will uniformly display the phenotype associated with the dominant allele.
- Law of Independent Assortment
- One of three fundamental principles of Mendelian inheritance, which states that genes responsible for different phenotypic traits are segregated independently during meiosis. Linked genes are a notable exception to this rule.
- Law of Segregation
- One of three fundamental principles of Mendelian inheritance, which states that during meiosis, the alleles of each gene segregate from each other such that each resulting gamete carries only one allele of each gene.
- leader sequence
- See 5' untranslated region.
- leading strand
- In DNA replication, the nascent strand for which both DNA polymerase's direction of synthesis and that of the overall chain elongation are toward the replication fork, i.e. both occur in the 5' to 3' direction, resulting in a single, continuous elongation process with few or no interruptions. By contrast, the other nascent strand, known as the lagging strand, is assembled in a discontinuous process involving the ligation of short DNA fragments which are synthesized in the opposite direction, away from the replication fork.[4]
- left splicing junction
- The boundary between the left end (by convention, the 5' end) of an intron and the right (3') end of an adjacent exon in a pre-mRNA transcript.
- leptonema
- In meiosis, the first of five substages of prophase I, following interphase and preceding zygonema. During leptonema, the replicated chromosomes condense from diffuse chromatin into long, thin strands that are much more visible within the nucleus.
- lethal equivalent value
- The average number of recessive deleterious genes existing in the heterozygous condition that is carried by a member of a population of diploid organisms, multiplied by the average probability that each such gene will cause premature lethality when homozygous. For example, an organism carrying eight recessive semilethal alleles, each of which produces only a 50% probability of premature death when homozygous, is said to carry a genetic burden of four "lethal equivalents".[4]
- lethal mutation
- Any mutation that results in the premature death of the organism carrying it. Recessive lethal mutations are fatal only to homozygotes, whereas dominant lethals are fatal even in heterozygotes.[4]
- leucine zipper
- ligase
- A class of enzymes which catalyze the joining of large molecules such as nucleic acids by forming one or more chemical bonds between them, typically C–C, C–O, C–S, or C–N bonds via condensation reactions. An example is DNA ligase, which catalyzes the formation of phosphodiester bonds between adjacent nucleotides on one or both strands of a DNA molecule, a reaction known as ligation.
- ligation
- lineage
- A linear evolutionary sequence connecting an ancestral cell, organism, or species to a particular descendant cell, organism, or species, including all intermediate organisms and spanning any number of generations; the direct progression of reproductive events (i.e. the line of descent) between two individuals, including vertically related individuals, e.g. parent(s) and offspring, but usually excluding horizontally related individuals who did not themselves directly contribute genetic material to any of the included individuals, e.g. siblings.
- linkage
- The tendency of DNA sequences which are physically near to each other on the same chromosome to be inherited together during meiosis. Because the physical distance between them is relatively small, the chance that any two nearby parts of a DNA sequence (often loci or genetic markers) will be separated on to different chromatids during chromosomal crossover is statistically very low; such loci are then said to be more linked than loci that are farther apart. Loci that exist on entirely different chromosomes are said to be perfectly unlinked. The standard unit for measuring genetic linkage is the centimorgan (cM).
- linkage disequilibrium
- linker DNA
- 1. A short, synthetic DNA duplex containing the recognition sequence for a particular restriction enzyme.[4] In molecular cloning, linkers are often deliberately included in recombinant molecules in order to make them easily modifiable by permitting cleavage and insertion of foreign sequences at precise locations. A segment of an engineered plasmid containing many such restriction sites is sometimes called a polylinker.
- 2. A section of chromosomal DNA connecting adjacent nucleosomes by binding to histone H1.[4]
- linking number
- The number of times that the two strands of a circular double-helical DNA molecule cross each other, equivalent to the twisting number (which measures the torsion of the double helix) plus the writhing number (which measures the degree of supercoiling). The linking number of a closed molecule cannot be changed without breaking and rejoining the strands. DNA molecules which are identical except for their linking numbers are known as topological isomers.[4]
- lncRNA
- See long non-coding RNA.
- locus
- A specific, fixed position on a chromosome where a particular gene or genetic marker resides.
- LOD score
- long arm
- In condensed chromosomes where the positioning of the centromere creates two segments or "arms" of unequal length, the longer of the two arms of a chromatid. Contrast short arm.
- long interspersed nuclear element (LINE)
- Any of a large family of non-LTR retrotransposons which together comprises one of the most widespread mobile genetic elements in eukaryotic genomes. Each LINE insertion is on average about 7,000 base pairs in length.
- long non-coding RNA (lncRNA)
- A class of non-coding RNA consisting of all transcripts of more than 200 nucleotides in length that are not translated. This limit distinguishes lncRNA from the numerous smaller non-coding RNAs such as microRNA. See also long intervening non-coding RNA.
- lyonization
- See X-inactivation.
See also
- Introduction to genetics
- Outline of genetics
- Outline of cell biology
- Glossary of biology
- Glossary of chemistry
- Glossary of evolutionary biology
References
- ^ "Talking Glossary of Genetic Terms". genome.gov. 8 October 2017. Retrieved 8 October 2017.
- ^ a b Ussery, David W. "DNA Structure: A-, B-, and Z-DNA Helix Families". Lyngby, Denmark: Danish Technical University.
{{cite web}}: Missing or empty|url=(help) - ^ a b c d e Klug, William S.; Cummings, Michael R. (1986). Concepts of Genetics (2nd ed.). Glenview, Ill.: Scott, Foresman and Company. ISBN 0-673-18680-6.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac King, Robert C.; Stansfield, William D.; Mulligan, Pamela K. (2006). A Dictionary of Genetics (7th ed.). Oxford: Oxford University Press. ISBN 978-0-19-530762-7.
- ^ a b Lewin, Benjamin (2003). Genes VIII. Upper Saddle River, NJ: Pearson Prentice Hall. ISBN 0-13-143981-2.
- ^ Nishikawa, S. (2007). "Reprogramming by the numbers". Nature Biotechnology. 25 (8): 877–878. doi:10.1038/nbt0807-877. PMID 17687365. S2CID 39773318.
- ^ a b c d e f g h i Rieger, Rigomar (1991). Glossary of Genetics: Classical and Molecular (5th ed.). Berlin: Springer-Verlag. ISBN 3540520546.
- ^ Priness, I.; Maimon, O.; Ben-Gal, I. (2007). "Evaluation of gene-expression clustering via mutual information distance measure". BMC Bioinformatics. 8: 111. doi:10.1186/1471-2105-8-111. PMC 1858704. PMID 17397530.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Functional Genomics Data Society – FGED Society".
- ^ Edgar, R; Domrachev, M; Lash, AE (1 January 2002). "Gene Expression Omnibus: NCBI gene expression and hybridization array data repository". Nucleic Acids Research. 30 (1): 207–10. doi:10.1093/nar/30.1.207. PMC 99122. PMID 11752295.
- ^ Barrett, T; Wilhite, SE; Ledoux, P; Evangelista, C; Kim, IF; Tomashevsky, M; Marshall, KA; Phillippy, KH; Sherman, PM; Holko, M; Yefanov, A; Lee, H; Zhang, N; Robertson, CL; Serova, N; Davis, S; Soboleva, A (January 2013). "NCBI GEO: archive for functional genomics data sets--update". Nucleic Acids Research. 41 (Database issue): D991-5. doi:10.1093/nar/gks1193. PMC 3531084. PMID 23193258.
- ^ Resta R, Biesecker BB, Bennett RL, Blum S, Hahn SE, Strecker MN, Williams JL (April 2006). "A new definition of Genetic Counseling: National Society of Genetic Counselors' Task Force report". Journal of Genetic Counseling. 15 (2): 77–83. doi:10.1007/s10897-005-9014-3. PMID 16761103.
- ^ Yakovchuk P, Protozanova E, Frank-Kamenetskii MD (2006). "Base-stacking and base-pairing contributions into thermal stability of the DNA double helix". Nucleic Acids Res. 34 (2): 564–74. doi:10.1093/nar/gkj454. PMC 1360284. PMID 16449200.
Further reading
- Budd, A. (2012). "Introduction to genome biology: features, processes, and structures". Evolutionary Genomics. Methods in Molecular Biology. Vol. 855. pp. 3–4. doi:10.1007/978-1-61779-582-4_1. ISBN 978-1-61779-581-7. PMID 22407704.
