Folding@home: Difference between revisions
→Biomedical significance: improved first couple paragraphs |
m journal citations to "holy grail of computational biology" |
||
| Line 36: | Line 36: | ||
[[Protein]]s are an essential component to many biological functions, and participate in virtually every process within cells. They often act as [[enzymes]], performing biochemical reactions including [[cell signaling]], transportation, [[Cell cycle#Regulation of eukaryotic cell cycle|cellular regulation]], and others. As structural elements, some proteins act as a type of [[cytoskeleton|skeleton for cells]], and as [[antibodies]] other proteins help the [[immune system]]. Before a protein can take on these roles, it must fold into a functional [[Protein tertiary structure|three-dimensional structure]], a processes that often occurs spontaneously and is strongly dependent on the protein's amino acid sequence. Understanding protein folding is thus critical to understanding what a protein does and how it works. However, proteins may misfold — that is, it folds down the wrong pathway and end up misshapen — and unless cellular mechanisms are capable of destroying or refolding them, they can subsequently [[Protein aggregation|aggregate]] and cause diseases. The tendency for this to occur is heavily dependent on the protein's chemical properties and the surrounding cellular environment, and laboratory experiments studying this process can be severely limited in scope and detail. This has led to the development of physics-based computational models that, when complementing experiments, seek to provide a more complete picture of protein folding, misfolding, and aggregation.<ref>{{cite journal | author = F. Chiti, N. Taddei, G. Ramponi, and C. M. Dobson | title = Rationalization of the effects of mutations on peptide and protein aggregation rates | journal = Nature | year = 2003 | volume = | issue = | pages = 805-808 | doi = | pmc = | pmid = 12917692}}</ref><ref name="taming folding complexity">{{cite journal | author = G. Bowman, V. Volez, and V. S. Pande | title = Taming the complexity of protein folding | journal = Current Opinion in Structural Biology | year = 2011 | volume = 21 | issue = 1 | pages = 4–11 | doi = 10.1016/j.sbi.2010.10.006 | pmc = 3042729 | pmid = 21081274}}</ref><ref name="from the TT to organism">{{cite journal | author = Leila M Luheshi, Damian Crowther, Christopher Dobson | title = Protein misfolding and disease: from the test tube to the organism | journal = Current Opinion in Chemical Biology | year = 2008 | volume = 12 | issue = 1 | pages = 25–31 | doi = 10.1016/j.cbpa.2008.02.011 | issn = | pmc = | pmid = 18295611}}</ref><ref name="PF then and now">{{cite journal | author = Yiwen Chen, Feng Ding, Huifen Nie, Adrian W. Serohijos, Shantanu Sharma, Kyle C. Wilcox, Shuangye Yin, Nikolay V. Dokholyan | title = Protein folding: Then and now | journal = Archives of Biochemistry and Biophysics | year = 2008 | volume = 469 | issue = 1 | pages = 4–19 | doi = 10.1016/j.abb.2007.05.014 | issn = | pmc = | pmid = }}</ref> |
[[Protein]]s are an essential component to many biological functions, and participate in virtually every process within cells. They often act as [[enzymes]], performing biochemical reactions including [[cell signaling]], transportation, [[Cell cycle#Regulation of eukaryotic cell cycle|cellular regulation]], and others. As structural elements, some proteins act as a type of [[cytoskeleton|skeleton for cells]], and as [[antibodies]] other proteins help the [[immune system]]. Before a protein can take on these roles, it must fold into a functional [[Protein tertiary structure|three-dimensional structure]], a processes that often occurs spontaneously and is strongly dependent on the protein's amino acid sequence. Understanding protein folding is thus critical to understanding what a protein does and how it works. However, proteins may misfold — that is, it folds down the wrong pathway and end up misshapen — and unless cellular mechanisms are capable of destroying or refolding them, they can subsequently [[Protein aggregation|aggregate]] and cause diseases. The tendency for this to occur is heavily dependent on the protein's chemical properties and the surrounding cellular environment, and laboratory experiments studying this process can be severely limited in scope and detail. This has led to the development of physics-based computational models that, when complementing experiments, seek to provide a more complete picture of protein folding, misfolding, and aggregation.<ref>{{cite journal | author = F. Chiti, N. Taddei, G. Ramponi, and C. M. Dobson | title = Rationalization of the effects of mutations on peptide and protein aggregation rates | journal = Nature | year = 2003 | volume = | issue = | pages = 805-808 | doi = | pmc = | pmid = 12917692}}</ref><ref name="taming folding complexity">{{cite journal | author = G. Bowman, V. Volez, and V. S. Pande | title = Taming the complexity of protein folding | journal = Current Opinion in Structural Biology | year = 2011 | volume = 21 | issue = 1 | pages = 4–11 | doi = 10.1016/j.sbi.2010.10.006 | pmc = 3042729 | pmid = 21081274}}</ref><ref name="from the TT to organism">{{cite journal | author = Leila M Luheshi, Damian Crowther, Christopher Dobson | title = Protein misfolding and disease: from the test tube to the organism | journal = Current Opinion in Chemical Biology | year = 2008 | volume = 12 | issue = 1 | pages = 25–31 | doi = 10.1016/j.cbpa.2008.02.011 | issn = | pmc = | pmid = 18295611}}</ref><ref name="PF then and now">{{cite journal | author = Yiwen Chen, Feng Ding, Huifen Nie, Adrian W. Serohijos, Shantanu Sharma, Kyle C. Wilcox, Shuangye Yin, Nikolay V. Dokholyan | title = Protein folding: Then and now | journal = Archives of Biochemistry and Biophysics | year = 2008 | volume = 469 | issue = 1 | pages = 4–19 | doi = 10.1016/j.abb.2007.05.014 | issn = | pmc = | pmid = }}</ref> |
||
Since the 1990s molecular dynamics simulations have been severely limited by computational power.<ref name="PF then and now"/> In 2001 simulations could only achieve nanosecond to single microsecond timescales, while experiments revealed millisecond folding events.<ref name="Sim of small alpha-helical protein folding"/><ref name="taming folding complexity"/> Due to the complexity of the protein's [[configuration space|conformation space]] and the timescales over which it folds, accurately simulating protein folding is significantly beyond the capabilities of a single modern computer and is considered a "holy grail" of [[computational biology]].<ref |
Since the 1990s molecular dynamics simulations have been severely limited by computational power.<ref name="PF then and now"/> In 2001 simulations could only achieve nanosecond to single microsecond timescales, while experiments revealed millisecond folding events.<ref name="Sim of small alpha-helical protein folding"/><ref name="taming folding complexity"/> Due to the complexity of the protein's [[configuration space|conformation space]] and the timescales over which it folds, accurately simulating protein folding is significantly beyond the capabilities of a single modern computer and is considered a "holy grail" of [[computational biology]].<ref>{{cite journal | author = Fabrizio Marinelli1,2, Fabio Pietrucci1, Alessandro Laio1*, Stefano Piana | title = A Kinetic Model of Trp-Cage Folding from Multiple Biased Molecular Dynamics Simulations | journal = PLoS Computational Biology | year = 2009 | volume = 8 | issue = 5 | pages = | doi = 10.1371/journal.pcbi.1000452 | pmid = }}</ref><ref>{{cite journal | author = Joseph Bordogna | title = The 21st Century Engineer | journal = IEEE Spectrum | year = 2001 | volume = 38 | issue = 1 | pages = 17 | doi = | pmid = | issn = | url = http://repository.upenn.edu/cgi/viewcontent.cgi?article=1004&context=ese_papers}}</ref><ref>{{cite web | url = http://www.astrobio.net/pressrelease/299/cutting-through-the-protein-knots | title = Cutting through the Protein Knots | publisher = Astrobiology Magazine | date = 2002-10-24 | accessdate = 2012-02-22}}</ref> [[Supercomputer]]s have attempted to successfully to address this problem, but are intrinsically expensive, typically shared between hundreds of different research groups, and strong molecular simulation [[scalability|scaling]] is difficult.<ref name="Atomistic subms protein folding">{{cite journal | author = Vijay S. Pande, Ian Baker, Jarrod Chapman, Sidney P. Elmer, Siraj Khaliq, Stefan M. Larson, Young Min Rhee, Michael R. Shirts, Christopher D. Snow, Eric J. Sorin, Bojan Zagrovic | title = Atomistic protein folding simulations on the submillisecond timescale using worldwide distributed computing | journal = Biopolymers | year = 2002 | volume = 68 | issue = 1 | pages = 91–109 | doi = 10.1002/bip.10219 | pmid = 12579582}}</ref> Without the use of customized hardware, straightforward computations on biologically and experimentally relevant timescales typically have exceptional difficulty for all but the most elementary of systems, which has prompted the use of simplified models which may be insufficient for a comprehensive view of protein folding.<ref name="taming folding complexity"/><ref name="how well can simulation predict">{{cite journal | author = C. D. Snow, E. J. Sorin, Y. M. Rhee, and V. S. Pande. | title = How well can simulation predict protein folding kinetics and thermodynamics? | journal = Annual Reviews of Biophysics | year = 2005 | volume = 34 | issue = | pages = 43–69 | doi = 10.1146/annurev.biophys.34.040204.144447 | pmid = 15869383}}</ref> Moreover, a limited number of long simulations are not sufficient for revealing the protein dynamics because protein folding is intrinsically statistical.<ref name="taming folding complexity"/> |
||
Folding@home simulation techniques rely on the behavior of proteins to spend a significant amount of the folding time "waiting" in various unique [[Protein conformation|conformational]] [[Statistical ensemble (mathematical physics)|states]], each a [[Gibbs free energy|free energy]] minima, before quickly transitioning to the next configuration.<ref name="Atomistic subms protein folding"/> Based on a small set of initial data, (from previous simulations or experiments) the Pande lab can identify and organize thousands of these states and build [[Hidden Markov model|Markov state models]]. These MSMs use short simulations to find the statistical chances and rates of transitions between these states and essentially serves as a map of the protein's [[energy landscape|free energy landscape]] and [[kinetic energy|kinetic]] and [[equilibrium thermodynamics]] properties.<ref name="taming folding complexity"/><ref name="PF then and now"/><ref name="Atomistic subms protein folding"/> Not only can this information be determined in parallel for each transition, but the transitions themselves can also be individually computed, and the overall protein folding process can then be reconstructed. This [[parallelization]] allows for a significant reduction in serial calculation time. Each MSM is also capable of revealing which transitions are [[statistical error|limiting]] the accuracy of the model, which allow for specific follow-up simulations to efficiently improve the thoroughness of the model. Using this adaptive sampling technique, the amount of time it takes to construct an accurate Markov State Model is inversely proportional to the number of parallel simulations run. This statistical kinetic model can be viewed at an arbitrary resolution, reveals [[measurement uncertainty]] and diversity of the folding pathways, (including how the protein can misfold) allows simulations at biologically relevant timescales, and has accurately compared to experiments.<ref name="Everything about MSMs"/><ref name="taming folding complexity"/><ref name="Atomistic subms protein folding"/> In 2010, the Pande lab used Markov state models to simulate the millisecond-folder NTL9 protein, a thousand times longer than previously achieved, even though each individual trajectories run on Folding@home were two orders of magnitude shorter.<ref name="Everything about MSMs"/><ref>{{cite journal | author = Vincent A. Voelz, Gregory R. Bowman, Kyle Beauchamp and Vijay S. Pande | title = Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1–39) | journal = Journal of the American Chemical Society | year = 2010 | volume = 132 | issue = 5 | pages = 1526–1528 | doi = 10.1021/ja9090353 | pmid = 20070076 | pmc = 2835335}}</ref> This was the first demonstration that MSMs are capable of statistically capturing folding events that could not be seen by conventional simulation methods.<ref name="taming folding complexity"/> For the instrumental development of the software used to automatically build these MSMs and for attaining quantitative agreement between theory and experiment, in 2010 Folding@home researcher Greg Bowman was awarded the [[Thomas Kuhn]] Paradigm Shift Award from the [[American Chemical Society]].<ref>{{cite web | url = https://simtk.org/project/xml/news.xml?group_id=357 | title = Greg Bowman awarded the 2010 Kuhn Paradigm Shift Award | publisher = SimTK: MSMBuilder | date = 2010-03-29 | accessdate = 2011-11-30}}</ref><ref>{{cite web | url = http://web2011.acscomp.org/awards/thomas-kuhn-paradigm-shift-award | title = Thomas Kuhn Paradigm Shift Award | publisher = ACS: Computers in Chemistry | accessdate = 2011-11-30}}</ref> |
Folding@home simulation techniques rely on the behavior of proteins to spend a significant amount of the folding time "waiting" in various unique [[Protein conformation|conformational]] [[Statistical ensemble (mathematical physics)|states]], each a [[Gibbs free energy|free energy]] minima, before quickly transitioning to the next configuration.<ref name="Atomistic subms protein folding"/> Based on a small set of initial data, (from previous simulations or experiments) the Pande lab can identify and organize thousands of these states and build [[Hidden Markov model|Markov state models]]. These MSMs use short simulations to find the statistical chances and rates of transitions between these states and essentially serves as a map of the protein's [[energy landscape|free energy landscape]] and [[kinetic energy|kinetic]] and [[equilibrium thermodynamics]] properties.<ref name="taming folding complexity"/><ref name="PF then and now"/><ref name="Atomistic subms protein folding"/> Not only can this information be determined in parallel for each transition, but the transitions themselves can also be individually computed, and the overall protein folding process can then be reconstructed. This [[parallelization]] allows for a significant reduction in serial calculation time. Each MSM is also capable of revealing which transitions are [[statistical error|limiting]] the accuracy of the model, which allow for specific follow-up simulations to efficiently improve the thoroughness of the model. Using this adaptive sampling technique, the amount of time it takes to construct an accurate Markov State Model is inversely proportional to the number of parallel simulations run. This statistical kinetic model can be viewed at an arbitrary resolution, reveals [[measurement uncertainty]] and diversity of the folding pathways, (including how the protein can misfold) allows simulations at biologically relevant timescales, and has accurately compared to experiments.<ref name="Everything about MSMs"/><ref name="taming folding complexity"/><ref name="Atomistic subms protein folding"/> In 2010, the Pande lab used Markov state models to simulate the millisecond-folder NTL9 protein, a thousand times longer than previously achieved, even though each individual trajectories run on Folding@home were two orders of magnitude shorter.<ref name="Everything about MSMs"/><ref>{{cite journal | author = Vincent A. Voelz, Gregory R. Bowman, Kyle Beauchamp and Vijay S. Pande | title = Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1–39) | journal = Journal of the American Chemical Society | year = 2010 | volume = 132 | issue = 5 | pages = 1526–1528 | doi = 10.1021/ja9090353 | pmid = 20070076 | pmc = 2835335}}</ref> This was the first demonstration that MSMs are capable of statistically capturing folding events that could not be seen by conventional simulation methods.<ref name="taming folding complexity"/> For the instrumental development of the software used to automatically build these MSMs and for attaining quantitative agreement between theory and experiment, in 2010 Folding@home researcher Greg Bowman was awarded the [[Thomas Kuhn]] Paradigm Shift Award from the [[American Chemical Society]].<ref>{{cite web | url = https://simtk.org/project/xml/news.xml?group_id=357 | title = Greg Bowman awarded the 2010 Kuhn Paradigm Shift Award | publisher = SimTK: MSMBuilder | date = 2010-03-29 | accessdate = 2011-11-30}}</ref><ref>{{cite web | url = http://web2011.acscomp.org/awards/thomas-kuhn-paradigm-shift-award | title = Thomas Kuhn Paradigm Shift Award | publisher = ACS: Computers in Chemistry | accessdate = 2011-11-30}}</ref> |
||
| Line 87: | Line 87: | ||
[[Image:FAH-tflops.PNG| thumb | left | 260px | Folding@home computing power shown, by device type, in tera[[FLOPS]] as recorded semi-daily from November 2006 until September 2007. Note the large spike in total compute power after March 22, when the [[PlayStation 3]] client was released.]] |
[[Image:FAH-tflops.PNG| thumb | left | 260px | Folding@home computing power shown, by device type, in tera[[FLOPS]] as recorded semi-daily from November 2006 until September 2007. Note the large spike in total compute power after March 22, when the [[PlayStation 3]] client was released.]] |
||
Interest and participation in the project has grown steadily since its launch.<ref>{{cite web | url = http://folding.typepad.com/news/2007/10/fun-fact-fah-gr.html | title = Fun fact: FAH growth over time | author = Vijay Pande | date = 2007-10-21 | accessdate = 2011-10-21}}</ref><ref>{{cite web | url = http://www.stanford.edu/group/pandegroup/images/ActiveCPUs2010.png | title = Active CPUs | format = Image | author = Pande lab | publisher = [[Stanford University]] | accessdate = 2011-08-30}}</ref> As of January 26, 2011, Folding@home has about 375,000 active [[CPU]]s, about 37,000 active [[GPU]]s, and about 22,000 active [[PlayStation 3|PS3s]], for a total of about 6.5 native [[petaflop|petaFLOPS]], (8.8 [[x86]] petaFLOPS) more computing power than the combined efforts of all distributed computing projects under BOINC.<ref name="osstats">{{cite web | url = http://fah-web.stanford.edu/cgi-bin/main.py?qtype=osstats | title = Client Statistics by OS | author = Pande lab | date = updated daily | publisher = Stanford University | accessdate = 2011-12-23}}</ref><ref>{{cite web | url = http://boincstats.com/stats/project_graph.php?pr=bo | title = BOINC Combined Credit Overview | accessdate = 2012-01-26}}</ref> A large majority of this performance comes from the GPU and PS3 clients.<ref name="osstats"/> With the Markov state model approach, Folding@home achieves [[strong scaling]] across its user base; it gains a near-linear speedup for every additional processor.<ref name="Atomistic subms protein folding"/><ref>{{cite journal | author = Michael R. Shirts and Vijay S. Pande | title = Mathematical Foundations of Coupled Parallel Simulations | journal = Physical Review Letters | year = 2001 | volume = 86 | issue = 22 | pages = 4983–4987 | doi = 10.1103/PhysRevLett.86.4983 | pmid = 11384401 | bibcode = 2001PhRvL..86.4983S}}</ref> In 2007, [[Guinness World Records|Guinness]] recognized Folding@home as the most powerful distributed computing cluster in the world.<ref>{{cite web | url = http://www.engadget.com/2007/10/31/folding-home-recognized-by-guinness-world-records/ | title = Folding@Home recognized by Guinness World Records | author = Joshua Topolsky | date = 2007-10-31 | accessdate = 2007-11-05}}</ref> This large and powerful network allows FAH to do work not possible any other way, including through the use of supercomputers,<ref>{{cite journal | author = Caroline Hadley | title = Biologists think bigger | journal = EMBO reports | year = 2004 | volume = 12 | issue = 5 | pages = 236–238 | doi = 10.1038/sj.embor.7400108 | pmid = | url = http://www.nature.com/embor/journal/v5/n3/full/7400108.html}}</ref> which are typically expensive to operate and often shared.<ref name="lessons from 8 years"/><ref name="Atomistic subms protein folding"/> |
Interest and participation in the project has grown steadily since its launch.<ref>{{cite web | url = http://folding.typepad.com/news/2007/10/fun-fact-fah-gr.html | title = Fun fact: FAH growth over time | author = Vijay Pande | date = 2007-10-21 | accessdate = 2011-10-21}}</ref><ref>{{cite web | url = http://www.stanford.edu/group/pandegroup/images/ActiveCPUs2010.png | title = Active CPUs | format = Image | author = Pande lab | publisher = [[Stanford University]] | accessdate = 2011-08-30}}</ref> As of January 26, 2011, Folding@home has about 375,000 active [[CPU]]s, about 37,000 active [[GPU]]s, and about 22,000 active [[PlayStation 3|PS3s]], for a total of about 6.5 native [[petaflop|petaFLOPS]], (8.8 [[x86]] petaFLOPS) more computing power than the combined efforts of all distributed computing projects under BOINC.<ref name="osstats">{{cite web | url = http://fah-web.stanford.edu/cgi-bin/main.py?qtype=osstats | title = Client Statistics by OS | author = Pande lab | date = updated daily | publisher = Stanford University | accessdate = 2011-12-23}}</ref><ref>{{cite web | url = http://boincstats.com/stats/project_graph.php?pr=bo | title = BOINC Combined Credit Overview | accessdate = 2012-01-26}}</ref> A large majority of this performance comes from the GPU and PS3 clients.<ref name="osstats"/> With the Markov state model approach, Folding@home achieves [[strong scaling]] across its user base; it gains a near-linear speedup for every additional processor.<ref name="Atomistic subms protein folding"/><ref>{{cite journal | author = Michael R. Shirts and Vijay S. Pande | title = Mathematical Foundations of Coupled Parallel Simulations | journal = Physical Review Letters | year = 2001 | volume = 86 | issue = 22 | pages = 4983–4987 | doi = 10.1103/PhysRevLett.86.4983 | pmid = 11384401 | bibcode = 2001PhRvL..86.4983S}}</ref> In 2007, [[Guinness World Records|Guinness]] recognized Folding@home as the most powerful distributed computing cluster in the world.<ref>{{cite web | url = http://www.engadget.com/2007/10/31/folding-home-recognized-by-guinness-world-records/ | title = Folding@Home recognized by Guinness World Records | author = Joshua Topolsky | date = 2007-10-31 | accessdate = 2007-11-05}}</ref> This large and powerful network allows FAH to do work not possible any other way, including through the use of supercomputers,<ref>{{cite journal | author = Caroline Hadley | title = Biologists think bigger | journal = EMBO reports | year = 2004 | volume = 12 | issue = 5 | pages = 236–238 | doi = 10.1038/sj.embor.7400108 | pmid = | url = http://www.nature.com/embor/journal/v5/n3/full/7400108.html}}</ref> which are typically expensive to operate and often shared.<ref name="lessons from 8 years">{{cite journal | author = Adam Beberg, Daniel Ensign, Guha Jayachandran, Siraj Khaliq, Vijay Pande | title = Folding@home: Lessons From Eight Years of Volunteer Distributed Computing | journal = Parallel & Distributed Processing, IEEE International Symposium | year = 2009 | volume = | issue = | pages = 1–8 | doi = 10.1109/IPDPS.2009.5160922 | pmid = | issn = 1530-2075 | url = http://www.hicomb.org/papers/HICOMB2009-13.pdf}}</ref><ref name="Atomistic subms protein folding"/> |
||
Folding@home gained popularity early in its history. In March 2002, [[Google]] co-founder [[Sergey Brin]] launched [[Google Compute]] as add-on for the [[Google Toolbar]].<ref>{{Cite news | url = http://news.cnet.com/2100-1001-867091.html | title = Google takes on supercomputing | date = March 22, 2002 | publisher = CNet News | first = Stephen | last = Shankland}}</ref> Although limited in functionality and scope, it increased Folding@home's participation from 10,000 up to about 30,000 active CPUs.<ref name="Biotech 27">{{cite web | url = http://castroller.com/Podcasts/FuturesInBiotech/249153 | title = Futures in Biotech 27: Folding@home at 1.3 Petaflops | format = Interview, webcast | date = 2007-12-28}}</ref> The program ended in October 2005 in favor of the Pande lab's official clients, and is no longer available for the Toolbar.<ref>{{Cite news | url = http://hardforum.com/showpost.php?p=1028770683&postcount=4 | title = Google is after your CPU cycles | author = ChelseaOilman | date = 2005-12-30 | accessdate = 2011-09-06}}</ref><ref>{{Cite news | url = http://toolbar.google.com/dc/offerdc.html/ | title = Your computer's idle time is too precious to waste | author = Google | year = 2007 | accessdate = 2011-09-06}}</ref> Folding@home also gained participants from [[Genome@home]], another distributed computing project from the Pande lab and a sister project to Folding@home. The goal of Genome@home was [[protein design]] and its applications, and the project was officially concluded in March 2004. Following its completion, users were asked to donate to Folding@home instead.<ref>{{Cite news | url = http://www.stanford.edu/group/pandegroup/genome/new.html | title = Genome@home Updates | date = 2002-03-04 | accessdate = 2011-09-05}}</ref><ref>{{Cite news | url = http://genomeathome.stanford.edu/faq.html | title = Genome@home FAQ | author = Pande lab | publisher = [[Stanford University]] | format = FAQ | accessdate = 2011-09-05}}</ref> |
Folding@home gained popularity early in its history. In March 2002, [[Google]] co-founder [[Sergey Brin]] launched [[Google Compute]] as add-on for the [[Google Toolbar]].<ref>{{Cite news | url = http://news.cnet.com/2100-1001-867091.html | title = Google takes on supercomputing | date = March 22, 2002 | publisher = CNet News | first = Stephen | last = Shankland}}</ref> Although limited in functionality and scope, it increased Folding@home's participation from 10,000 up to about 30,000 active CPUs.<ref name="Biotech 27">{{cite web | url = http://castroller.com/Podcasts/FuturesInBiotech/249153 | title = Futures in Biotech 27: Folding@home at 1.3 Petaflops | format = Interview, webcast | date = 2007-12-28}}</ref> The program ended in October 2005 in favor of the Pande lab's official clients, and is no longer available for the Toolbar.<ref>{{Cite news | url = http://hardforum.com/showpost.php?p=1028770683&postcount=4 | title = Google is after your CPU cycles | author = ChelseaOilman | date = 2005-12-30 | accessdate = 2011-09-06}}</ref><ref>{{Cite news | url = http://toolbar.google.com/dc/offerdc.html/ | title = Your computer's idle time is too precious to waste | author = Google | year = 2007 | accessdate = 2011-09-06}}</ref> Folding@home also gained participants from [[Genome@home]], another distributed computing project from the Pande lab and a sister project to Folding@home. The goal of Genome@home was [[protein design]] and its applications, and the project was officially concluded in March 2004. Following its completion, users were asked to donate to Folding@home instead.<ref>{{Cite news | url = http://www.stanford.edu/group/pandegroup/genome/new.html | title = Genome@home Updates | date = 2002-03-04 | accessdate = 2011-09-05}}</ref><ref>{{Cite news | url = http://genomeathome.stanford.edu/faq.html | title = Genome@home FAQ | author = Pande lab | publisher = [[Stanford University]] | format = FAQ | accessdate = 2011-09-05}}</ref> |
||
Revision as of 05:24, 23 February 2012
 | |
| Original author(s) | Vijay Pande |
|---|---|
| Developer(s) | Stanford University / Pande lab |
| Initial release | 2000-10-01 |
| Stable release | |
| Preview release | |
| Operating system | Microsoft Windows, Mac OS X, Linux |
| Platform | Cross-platform |
| Available in | English |
| Type | Distributed computing |
| License | Proprietary[6] |
| Website | folding |
Folding@home is a distributed computing project designed to use spare processing power on personal computers to perform simulations of disease-relevant protein folding and other molecular dynamics, and to improve on the methods of doing so. Also referred to as FAH or F@h, much of its work attempts to determine how proteins reach their final structure, which is of significant academic interest and has major implications to disease research. To a lesser degree Folding@home also tries to predict that final structure from only the initial amino acid sequence, which has applications in drug design.[7][8] Folding@home is developed and operated by the Pande Laboratory at Stanford University, under the direction of Vijay Pande. The goal of the project is to "understand protein folding, misfolding, and related diseases".[9][10]
Folding@home's simulations of protein folding and misfolding enable the scientific community to better understand the development of many diseases, including Alzheimer's disease, Parkinson's disease, cancer, Creutzfeldt–Jakob disease, Huntington's disease, cystic fibrosis, sickle-cell anaemia, HIV, Chagas disease, influenza, osteogenesis imperfecta, autism,[11] and alpha 1-antitrypsin deficiency, among others.[12] More fundamentally, understanding the process of protein folding — how proteins assemble themselves into a functional state — is one of the outstanding problems of molecular biology.[13] Since the project's launch on October 1st 2000,[9] the Pande lab has produced ninety-five scientific research papers as a direct result of the project using simulation methodology that is a paradigm shift away from traditional computational approaches.[14][15] These techniques have demonstrated accuracy compared to results from laboratory research, a "grand challenge" in computational biology.
Folding@home pioneered the uses of GPUs, PlayStation 3s, and Message Passing Interface (designed for multi-core processors) for distributed computing. In January 2010 the Folding@home project successfully simulated protein folding in the 1.5 millisecond range — a simulation thousands of times longer than previously achieved. It remains one of the world's fastest computing systems, and is more powerful than all distributed computing projects under BOINC combined. The Pande lab collaborates with and shares this resource with various scientific institutions and laboratories across the world.[16]
Biomedical significance
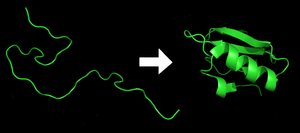
Proteins are an essential component to many biological functions, and participate in virtually every process within cells. They often act as enzymes, performing biochemical reactions including cell signaling, transportation, cellular regulation, and others. As structural elements, some proteins act as a type of skeleton for cells, and as antibodies other proteins help the immune system. Before a protein can take on these roles, it must fold into a functional three-dimensional structure, a processes that often occurs spontaneously and is strongly dependent on the protein's amino acid sequence. Understanding protein folding is thus critical to understanding what a protein does and how it works. However, proteins may misfold — that is, it folds down the wrong pathway and end up misshapen — and unless cellular mechanisms are capable of destroying or refolding them, they can subsequently aggregate and cause diseases. The tendency for this to occur is heavily dependent on the protein's chemical properties and the surrounding cellular environment, and laboratory experiments studying this process can be severely limited in scope and detail. This has led to the development of physics-based computational models that, when complementing experiments, seek to provide a more complete picture of protein folding, misfolding, and aggregation.[17][18][19][20]
Since the 1990s molecular dynamics simulations have been severely limited by computational power.[20] In 2001 simulations could only achieve nanosecond to single microsecond timescales, while experiments revealed millisecond folding events.[8][18] Due to the complexity of the protein's conformation space and the timescales over which it folds, accurately simulating protein folding is significantly beyond the capabilities of a single modern computer and is considered a "holy grail" of computational biology.[21][22][23] Supercomputers have attempted to successfully to address this problem, but are intrinsically expensive, typically shared between hundreds of different research groups, and strong molecular simulation scaling is difficult.[24] Without the use of customized hardware, straightforward computations on biologically and experimentally relevant timescales typically have exceptional difficulty for all but the most elementary of systems, which has prompted the use of simplified models which may be insufficient for a comprehensive view of protein folding.[18][25] Moreover, a limited number of long simulations are not sufficient for revealing the protein dynamics because protein folding is intrinsically statistical.[18]
Folding@home simulation techniques rely on the behavior of proteins to spend a significant amount of the folding time "waiting" in various unique conformational states, each a free energy minima, before quickly transitioning to the next configuration.[24] Based on a small set of initial data, (from previous simulations or experiments) the Pande lab can identify and organize thousands of these states and build Markov state models. These MSMs use short simulations to find the statistical chances and rates of transitions between these states and essentially serves as a map of the protein's free energy landscape and kinetic and equilibrium thermodynamics properties.[18][20][24] Not only can this information be determined in parallel for each transition, but the transitions themselves can also be individually computed, and the overall protein folding process can then be reconstructed. This parallelization allows for a significant reduction in serial calculation time. Each MSM is also capable of revealing which transitions are limiting the accuracy of the model, which allow for specific follow-up simulations to efficiently improve the thoroughness of the model. Using this adaptive sampling technique, the amount of time it takes to construct an accurate Markov State Model is inversely proportional to the number of parallel simulations run. This statistical kinetic model can be viewed at an arbitrary resolution, reveals measurement uncertainty and diversity of the folding pathways, (including how the protein can misfold) allows simulations at biologically relevant timescales, and has accurately compared to experiments.[15][18][24] In 2010, the Pande lab used Markov state models to simulate the millisecond-folder NTL9 protein, a thousand times longer than previously achieved, even though each individual trajectories run on Folding@home were two orders of magnitude shorter.[15][26] This was the first demonstration that MSMs are capable of statistically capturing folding events that could not be seen by conventional simulation methods.[18] For the instrumental development of the software used to automatically build these MSMs and for attaining quantitative agreement between theory and experiment, in 2010 Folding@home researcher Greg Bowman was awarded the Thomas Kuhn Paradigm Shift Award from the American Chemical Society.[27][28]
The Pande lab and other researchers can use Folding@home to study aspects of folding, misfolding, and related diseases that would never be seen experimentally.[29] For example, most proteins have such intrinsically stable native states that it is difficult to experimentally study their folding events, and researchers must resort to chemical denaturation methods.[30] While the denatured state of proteins is of scientific interest because any residual structure may affect its folding behavior, even with molecular experiments it remains difficult to determine the extent of a local structure that may be present. However, molecular simulations on Folding@home can provide detail into denatured conformational states and insights into the chemical denaturation mechanisms.[30] These and other simulations run on Folding@home are used in conjunction with laboratory experiments.[29] Additionally, as of 2011 the Pande lab is performing studies into how protein folding in their native cells may be different than in environments such as test tubes used during experiments.[31] Protein folding often occurs in crowded cellular environments, such as in the ribosome's exit tunnel or inside protein chaperones.[32][33]
In addition to the diseases listed below, researchers have used Folding@home to study malaria, Chagas disease, the misfolding of alpha-synuclein that is linked to Parkinson's disease, the amylin peptide involved in type II diabetes,[34] and the prions which cause Creutzfeldt–Jakob disease.[12][35] Results from this research have led to major shifts in the understanding of protein folding and its applications for disease, as well as improved protein folding models.[36][37] Folding@home is dedicated to producing significant amounts of results towards protein folding, the diseases that result from protein misfolding, and novel computational methods for doing so.[12] The goal of the first five years of the project was to make significant advances in understanding folding, while the current goal is to understand misfolding and related disease, especially Alzheimer's disease.[29] In 2002, Folding@home completed approximately a million CPU days of simulations over the span of several months.[38]
As a part of Stanford University, a non-profit organization, the Pande lab does not sell the results generated by Folding@home. The large data sets from the project are freely available for other researchers to use upon request, and some can be accessed from the Folding@home website.[39][40] The Pande lab also releases Folding@home's key software to other researchers, so that the algorithms which benefit Folding@home will also aid other scientific areas.[39] Moreover, in 2011 they released the open-source Copernicus software, so that other researchers can run molecular simulations much more efficiently on clusters or supercomputers.[41] Summaries of all of the scientific findings from Folding@home are posted on the Folding@home website after publication.[14] The full publications are available online or from a local municipal or academic library.[40]
Alzheimer's disease

Alzheimer's disease, a form of dementia which most often affects the elderly, is believed to be caused by specific misfolding and subsequent aggregation of the small 42-residue amyloid beta (Aß) peptide. The severity of the disease depends not only on the amount of Aß, but also on how it misfolds. Current theory holds that toxic non-plaque Aß oligomers (aggregates of many monomers) bind to a surface receptor on neurons and change the structure of the synapse, thereby disrupting neuronal communication and causing neuronal cell death which leads to the associated neurodegenerative consequences.[42] In 2011, Folding@home completed simulations of several mutant forms of this peptide, the results of which could aid in the development of therapeutic approaches to AD,[43] and will also help the Pande lab prepare for similar aggregation studies.[44]
Despite this connection to the disease, toxic Aß aggregations remain so complex that it was not previously possible to simulate them in atomic resolution. In 2011, the Pande lab explored how their Abeta studies using Folding@home could be used as a starting point for a new Alzheimer's therapy. Folding@home is currently concentrating on Alzheimer's and continues full-scale simulations of amyloid beta and its oligomerization,[45][46] which had previously been a technological challenge to simulate.[44] These studies build off of the Pande lab's 2008 published work into new ways to simulate Abeta oligomerization over long timescales. In the same publication, previous all-atom simulations were performed that led to specific experimentally-tested predictions, such as ways in which to stabilize the protein and prevent the toxic oligomer formation. The Pande lab is focusing their research in this area for rational drug design approaches.[12] Pande described that paper as the "tip of the iceberg" for the Folding@home studies of Alzheimer's, as further results will follow and possibly new therapeutics.[44][45]
Folding@home is also being used to study Aß fragments of different sizes to determine how various natural enzymes affect the structure and folding of Aß. These fragments are tied to senile plaques, a pathological marker of Alzheimer's disease in patient's brain. When certain enzymes cleave the amyloid precursor protein, Abeta peptides are produced, while the action of other enzymes can instead produce p3 peptides, much smaller fragments of Aß. Folding@home is simulating one of these smaller peptides in water in an effort to determine how the length of Aß affects its overall structure.[47]
In 2010, several possible drug leads predicted by Folding@home went from the test tube to testing on living tissue, and in close cooperation with the Nanomedicine Center for Protein Folding, the drug leads continued to be refined. Additionally, as predicted by FAH's simulations, a stable form of amyloid beta was experimentally verified which the Pande lab believes could be used as a starting point for new Alzheimer's therapy.[44][45] In 2008, Folding@home produced several small drug candidates to fight Alzheimer's Disease, as they appear to inhibit the toxicity of Abeta.[48]
The Pande lab is also using Folding@home to investigate protein–protein interactions, which occur extensively throughout both benign and disease-related biological activities. Interactions involving the common SH3 protein are also being studied, as it has implications in Alzheimers research. The refinement of these simulations has greatly improved the Pande lab's ability to understand a wide variety of biological interactions.[49]
Huntington's disease
Huntington's disease, an incurable neurodegenerative genetic disorder affecting muscle coordination and leading to dementia, is also associated with protein misfolding. Specifically, it is caused by a mutation in the Huntingtin gene, which causes excessively long repetitive chains of the glutamine amino acid in the Huntingtin protein, a protein that plays important roles in nerve cells.[50][51] The likelihood of neuronal cell death is primarily affected by the length of the glutamine chain and the neuron's intracellular exposure to the misfolded Huntingtin protein.[52] The defective protein causes Huntington's by aggregating most often in the striatum and frontal cortex of patient's brains. The Pande lab is using Folding@home to study these aggregates, as well as predict how they form.[12] How this aggregation occurs has been largely unknown, but in 2009 a paper based on Folding@home's results and published in the Journal of Molecular Biology investigated possible mechanisms for the aggregation formation, and the implications into how to prevent it.[51] These studies will be useful for drug design approaches against the disease, and will serve as a foundation for methods to stop the aggregation formation. Additionally, some of the methods used to study Huntington's are also being used for Alzheimer's research.[12]
In 2010, Folding@home researcher Veena Thomas proposed a novel therapeutic strategy for HD, which may be funded by the NIH. This strategy could be used to bring the results from Folding@home directly to a therapeutic.[12]
Cancer
More than half of all known cancers involve mutations of p53, a tumor suppressor protein present in every cell which signals for cell death in the event of damage to a cell's DNA. Specific mutations can cause p53 to misfold and become unable to send the "stop signal", consequently causing unchecked cell division. The type of mutation differs among the different locations of cancer, and analysis of these mutations is important for understanding the cause of these tumors.[53] In 2006 and in agreement with experimental data, the Pande lab's novel computational methods demonstrated reasonable success in reliably and rapidly identifying specific mutations that are linked to cancer. Moreover, the study was able to determine the effects of mutations that could not be experimentally measured. Further analysis continues of all possible amino acid possibilities at each position, in order to thoroughly predict cancer-associated mutations.[54] This followed work in 2005 which studied the dimerization of the p53 oligomerization domain, was the first study to examine the refolding of a protein dimer using molecular simulations in explicit water, and was the first peer-reviewed publication from a distributed computing project related to cancer.[55][56] Following these results, the Pande lab have expanded their efforts to other p53-related diseases.[12]
The Pande lab is also performing research into protein chaperones. These are proteins that assist in the folding of other molecules, assembly of oligomeric structures, the prevention of potential damage caused by protein misfolding, and other functions. They are needed for these purposes by rapidly growing cancerous cells.[57] Using Folding@home and working closely with the Protein Folding Center, they plan to find ways to inhibit chaperones involved in cancer. Using Folding@home for a more comprehensive visualization of their functions, the Pande lab and the Protein Folding Center collectively plan to engineer modified chaperonins to inhibit the folding of particular proteins associated with human diseases such as cancer and Alzheimer's. While this approach has been used before, they believe that this project, if successful, could lead to an interesting new drug against cancer or at least make major advances in that area.[57]
Folding@home is also used to study the folding of several other proteins which have mutations tied to cancer, such as the enzyme src Kinase and certain forms of the Engrailed homeodomain. These proteins also have a great deal of experimental data for comparison, and serve as a great system for the understanding of folding and misfolding.[58][59] Additionally, the Pande lab is using Folding@home to understand the dynamics of a small knottin protein and how it can be used to bind to contrast agents for imaging scan or drugs.[60] Finally, some forms of interleukin-2, an important signaling protein for the immune system, have been used as immunotherapy for cancer. The Pande lab believes that Folding@home's simulations of its dynamics will lead to insights into how to design other therapeutics.[61]
Osteogenesis imperfecta
Osteogenesis imperfecta is a non-curable genetic bone disorder. Those with the disease are unable to successfully make functional connective bone tissue. This is lethal for many but can also induce a higher rate of miscarriages.[12] The disease is caused by mutations in the Type-1 collagen protein, the most common form of collagen and found abundantly throughout the body. Although some of these mutations of collagen can lead to serious morphological disorders, more benign forms can cause brittle bones and other subtleties.[12] Folding@home has performed simulations of collagen, and has produced a paper on Osteogenesis imperfecta outlining new molecular simulation techniques and revealing new insights into how collagen misfolds. The Pande lab believes these results will be useful for later computational studies of collagen.[62]
Viruses
Folding@home is also performing simulations of certain viruses such as influenza and HIV. These viruses are widespread, have a history of causing global pandemics, and are exceptionally difficult to treat. While the majority of treatments focus on preventing viral replication, Folding@home's research concentrates on preventing the virus from entering cellular membranes in the first place.[12] Membrane fusion is a primary action that occurs during this viral entry, and an understanding of this process has implications for antiviral drugs and national health. Unfortunately, membrane mechanisms are difficult to analyse experimentally. Molecular dynamics simulations run on Folding@home can simulate the protein-lipid interactions involved during cellular viral infection, which have improved the Pande lab's understanding of the molecular dynamics behind this process.[12][63] Research using Folding@home also continues on the dynamics of the enzyme RNase H, a key component of HIV, in the hopes of designing drugs to deactivate it.[64]
Drug design
Once it is determined how a protein misfolds, preventative treatments can be applied during the folding process.[20] The combination of computational molecular modeling and experimental analysis has the possibility to fundamentally shape the future of molecular medicine and the rational design of therapeutics.[19] Computational drug design has two components; protein docking and free energy calculation. These are difficult and time-consuming to determine experimentally and traditional computational approaches must usually trade speed for accuracy.[65] Moreover, free energy calculation of molecular processes is a challenge in computational chemistry, but is pharmaceutically useful.[66][67] In 2006 the Pande lab presented methods for FAH which parallelized these calculations, which increased significantly increased computational efficiency compared to previous techniques. These methods had implications in docking, molecular dynamics, and free energy perturbation,[66] and allowed Folding@home to perform drug design calculations that were otherwise infeasible.[29][67]
Scientists can utilize Folding@home to precisely demonstrate how potential drugs will bind to proteins,[65] which is important because a ligand that strongly binds to a target protein is a promising drug candidate.[67] In 2011 as part of a blind experiment, Folding@home attempted to predict which of a set of ligands would attach to a target protein and also estimate their associated binding energies.[68][69] As of 2011, Folding@home is searching for prime binding locations on protein surfaces by testing the interactions of different molecules with known binding sites.[70] Additionally, studies are also underway on the dynamics of beta-lactamase, a protein that plays important roles in drug resistance. Following the results from this study, the Pande lab may be able to design drugs to deactivate it.[71]
As part of the drug design efforts, Folding@home also simulates the ribosome, a large biological machine that synthesizes proteins from mRNA. It is targeted by approximately half of all known antibiotics, which usually kill bacteria by preventing their ribosomes from making new and essential proteins. The full structure of the ribosome has only been recently determined, but the functions of many ribosomal proteins remain largely unknown.[72] In 2007 the Pande lab received a grant to study and design new antibiotics. The size and complexity of the ribosome has prepared the Pande lab for more complex biomedical problems.[12]
Participation

Interest and participation in the project has grown steadily since its launch.[73][74] As of January 26, 2011, Folding@home has about 375,000 active CPUs, about 37,000 active GPUs, and about 22,000 active PS3s, for a total of about 6.5 native petaFLOPS, (8.8 x86 petaFLOPS) more computing power than the combined efforts of all distributed computing projects under BOINC.[75][76] A large majority of this performance comes from the GPU and PS3 clients.[75] With the Markov state model approach, Folding@home achieves strong scaling across its user base; it gains a near-linear speedup for every additional processor.[24][77] In 2007, Guinness recognized Folding@home as the most powerful distributed computing cluster in the world.[78] This large and powerful network allows FAH to do work not possible any other way, including through the use of supercomputers,[79] which are typically expensive to operate and often shared.[80][24]
Folding@home gained popularity early in its history. In March 2002, Google co-founder Sergey Brin launched Google Compute as add-on for the Google Toolbar.[81] Although limited in functionality and scope, it increased Folding@home's participation from 10,000 up to about 30,000 active CPUs.[82] The program ended in October 2005 in favor of the Pande lab's official clients, and is no longer available for the Toolbar.[83][84] Folding@home also gained participants from Genome@home, another distributed computing project from the Pande lab and a sister project to Folding@home. The goal of Genome@home was protein design and its applications, and the project was officially concluded in March 2004. Following its completion, users were asked to donate to Folding@home instead.[85][86]
PetaFLOPS milestones
| Native petaFLOPS threshold | Date crossed | Fastest Supercomputer at Date CrossedNote 1 |
|---|---|---|
| 1.0 | September 16, 2007 | 0.2806 petaFLOP BlueGene/L[87] |
| 2.0 | May 7, 2008 | 0.4782 petaFLOP BlueGene/L[88] |
| 3.0 | August 20, 2008 | 1.042 petaFLOP Roadrunner[89] |
| 4.0 | September 28, 2008 | 1.042 petaFLOP Roadrunner[89] |
| 5.0 | February 18, 2009 | 1.105 petaFLOP Roadrunner[90] |
| 6.0 | November 10, 2011 | 8.162 petaFLOP K computer[91] |
On September 16, 2007, the Folding@home project officially attained a sustained performance level higher than one native petaFLOPS, becoming the first computing system of any kind in the world to do so,[92][93] although it had erroneously almost reached that level in March of that year.[94][95] On May 7, 2008, the project attained a sustained performance level higher than two native petaFLOPS,[96] followed by the three and four native petaFLOPS milestones on August 20 and September 28, 2008 respectively.[97][98] Then on February 18, 2009, Folding@home achieved a performance level of just above five native petaFLOPS, thereby becoming the first computing system to surpass that performance, just as it was for the other four milestones.[99][100] Most recently, on November 10, 2011, Folding@home crossed the six native petaFLOP barrier with the equivalent of nearly eight x86 petaFLOPS.[101]
Starting in March 2009, Folding@home began reporting performance in both native and x86 FLOPS.[102] While native FLOPS are a measure of the performance from a given hardware, Folding@home also estimates how many FLOPS the calculation would take on the standard x86 CPU architecture, which is commonly used as a performance reference. For instance, certain complex functions can be performed in one native FLOP on a GPU, but take multiple FLOPS on the x86 architecture.[103] Despite using conservative conversions based on the actual execution time of the calculations, for the GPU and PS3 clients x86 FLOPS are consistently much greater than the native FLOPS.[75][104] By reporting in both native and x86 FLOPS, Folding@home attempts to even out these hardware differences.[103]
Points
Distributed computing projects such as Folding@home are often driven by a sense of collegiate competition to compute the most for the project. Folding@home quantitatively assesses this through a point system.[105][106] Donors are granted point credit as a measure of their contribution, and these points can foster friendly competition between donors.[80] Points are determined by the performance of each contributor's folding hardware relative to a reference machine, and one or more Work Units from a project are benchmarked on that machine before the project is released. As some simulations are exceptionally demanding on a system, or are of great scientific priority, donors who opt-in and reliably complete these Work Units are non-linearly rewarded additional bonus points.[106][107] This generates a fair system of equal pay for equal work, and attempts to align credit with the value of the scientific results.[105][108] Donors can also use a passkey to securely protect their contributions, as they not only allow for the receipt of bonus points, but they also separate a donor from any policy issues arising from another using that username.[109][110]
Users can register their contributions under a team, which register the combined score of all their members. A user can start their own team, or they can join an existing team.[1][111] They can be used for troubleshooting or recruitment purposes, but can also keep donors motivated.[112] In some cases, a team may have their own community-driven sources of help such as a forum.[113][114] In addition, rivalries between teams create friendly competition that benefits the folding community,[115] and members can also have intra-team competitions for top spots.[116][117] However, regardless of username or team affiliation, all contributions go to the same place and have the same scientific value.[113] Rankings and other statistics for both individuals and teams are posted to the Folding@home website, with third party statistics sites also available.[118]
Software
Folding@home software on the user's end consists of three components: a client, work units, and cores.
Client
Folding@home is powered by volunteers who have installed a client program on their personal computer.[29] The project offers clients for multi-core processors, graphics processing units, and PlayStation 3s, complementing the standard client designed for uniprocessor systems. While these former clients use significantly more system resources, they also have the capability of completing an overall simulation very quickly, (in a few weeks or months rather than years) which is of major scientific value.[119] Folding@home is the first project to fully utilize GPUs,[120][121] PS3s,[122] and MPI for distributed computing.[123] As its software is custom-tailored to each hardware architecture, Folding@home gains the ability to run many different types of calculations, allowing the Pande lab to address biomedical questions previously considered impossible to tackle computationally.[119]
Each client is the software with which the user interacts, and manages the other software components behind the scenes.[124] Through the client, the user may pause the folding process, open an event log, check the work progress, or view personal statistics.[124] These clients run continuously in the background, using otherwise unused processing power.[1] These clients are designed to run FAH's calculations at an extremely low priority, and will back off to allow other computer programs to have more processing power.[125][126] Although modern computer chips are designed to be able to operate continuously without degrading,[125][127] if users wish to reduce power consumption or heat production, the maximum percentage of CPU power used can be adjusted if desired.[124] If interrupted by a computer shutdown or other means, the client will resume work at almost the same point at startup.[128] For users with machines with multiple processor units, multiple clients may be installed on one machine, and users may be credited by clients on multiple machines.[129]
For security and scientific integrity reasons, the Pande lab does not publicly release the source code of the clients.[6][130] Significant work goes into minimizing security issues in all of Folding@home's software.[129][131] For example, clients can be downloaded only from the official Folding@home website or its commercial partners.[6] It will upload and download data only from Stanford's Folding@home data servers, (over port 8080, with 80 as an alternative)[132] and will only interact with FAH computer files.[129][131] Moreover, it does not normally need computer administrative privileges,[131] so from a security standpoint it behaves similar to but is even more secure than a web browser.[82][132]
Folding@home's first client was a screensaver, which would run Folding@home while the computer was not otherwise in use.[124][133] Later, the Pande lab tested clients on the open source BOINC framework; however, this approach became unworkable and was abandoned in June 2006.[134] BOINC's fixed architecture limits the types of project it can accommodate and is not appropriate for Folding@home.[80]
Graphics processing units
Folding@home also utilizes GPUs for distributed computing. Their specialized hardware is designed to accelerate rendering of 3D graphics applications such as video games and can significantly outperform CPUs for certain types of calculations. Although limited in generality, this makes GPUs one of the most powerful, cost-effective, and rapidly growing computational platforms. As such, general purpose GPU computing is the pursuit of many scientists and researchers. However, GPU hardware is difficult to utilize for non-graphics task and usually requires significant algorithm restructuring and an advanced understanding of the underlying architecture.[135] Such custom-tailoring is challenging, especially to researchers with limited software development resources. To achieve hardware-independence, the Pande lab's open source OpenMM library serves as an high-level API, allowing molecular simulation software to run efficiently on varying architectures without significant modification. Lower-level APIs interface the higher-level API with the underlying platform. This flexible approach delivers performance nearly equal to hand-tuned GPU code, and greatly outperforms CPU implementations.[136][137] GPUs remain Folding@home's most powerful platform in terms of FLOPS; as of February 2012, GPU clients account for 76% of the entire project's x86 FLOP throughput.[75]
Up until 2010, the computational reliability of GPGPU consumer-grade hardware had remained largely unknown, and circumstantial evidence related to the lack of built-in error detection and correction in GPU memory raised reliability concerns. The Pande lab then conducted the first large-scale test of GPU scientific accuracy on over 20,000 hosts on the Folding@home network. Small memory soft errors were detected in two-thirds of tested GPUs. The study found that the error rate was most dependent on board architecture, but concluded that reliable GPGPU computing was very feasible as long as attention is paid to the hardware characteristics.[138]
The first generation of Folding@home's Windows GPU client (GPU1) was released to the public on October 2, 2006,[134] delivering a 20-30X speedup for certain calculations over its CPU-based Gromacs counterparts.[139] It was the first time GPUs had been used for either distributed computing or major molecular dynamics calculations.[120] Pande lab gained significant knowledge and experience with the development of GPGPU software, but citing a need to improve scientific accuracies over DirectX,[139][140] it was succeeded by GPU2, the second generation successor of the client on April 10, 2008.[141] Following its introduction, GPU1 was officially retired on June 6.[142] Compared to GPU1, GPU2 was more scientifically reliable and productive, ran on ATI and CUDA-enabled Nvidia GPUs, and supported more advanced algorithms, larger proteins, and real-time visualization of the protein simulation.[143][144] Following this, the third generation of Folding@home's GPU client (GPU3) was released on May 25, 2010. While backwards compatible to GPU2, GPU3 is comparatively more stable and efficient, has additional scientific capabilities,[145][146] and uses OpenMM on top of an OpenCL framework.[146][147] Although it does not natively support the Linux operating system, it can be run under WINE for donors with Nvidia graphics cards.[148][149]
PlayStation 3

Folding@home can also take advantage of the computing power of PlayStation 3s, to achieve performance previously only possible on supercomputers. Unlike Microsoft's Xbox 360, the PS3 is well suited for Folding@home simulations.[104] At the time of its inception and for certain calculations, its main streaming Cell processor delivered a 20x speed increase over PCs,[82][29] allowing the Pande lab to run calculations that would otherwise be computationally infeasible.[150] This high speed and efficiency introduced other opportunities for worthwhile optimizations, and radically changed the tradeoff between computational efficiency and overall accuracy, allowing for the utilization of more complex molecular models at little extra computational cost.[151] These capabilities allow for greater insights into disease research.[104]
The PS3 client was originally a standalone application, but since September 18, 2008 is a channel of Life with PlayStation, developed in a collaborative effort between Sony and the Pande lab.[104] It takes the middle path between a CPU's flexibility and a GPU's speed, performing a limited set of calculations rapidly while still retaining adaptable.[134] However, unlike CPUs and GPUs, donors cannot perform other activities on their PS3 while running Folding@home.[150] Instead, the Pande lab has specifically designed its Work Units to take approximately eight hours so that they can be completed overnight.[104] The PS3's uniform console environment makes support easier, as well as making Folding@home user friendly.[29] The PS3 also has the ability to stream data quickly to its GPU, allowing for real-time atomic detail visualizations of the protein dynamics.[151]
Multi-core processing client
The Symmetric MultiProcessing (SMP) client fulfills two purposes: it takes advantage of the high-performance capabilities of recent multiprocessor systems, and it helps develop a simulation architecture that will become one of the dominant FAH computing paradigms as multi-core chips become an industry standard over the next several years.[82][119][134] The SMP client is capable of delivering over a 4x calculation speedup over the standard uniprocessor clients.[119]
Folding@home's SMP core handles multi-core CPUs very different from other distributed computing projects, including those under BOINC.[152] Instead of simply doing multiple Work Units simultaneously, single WUs are completed much faster across the multiple CPU cores.[153] This cuts down on the traditional difficulties of scaling a large simulation to many processors. As such, this approach is very scientifically valuable. Some of the Pande lab publications would not have been possible without the SMP client.[153]
In November 2006, first generation SMP Folding@home clients for x86 Microsoft Windows, x86-64 Linux, and x86 Mac OS X were released.[134] These clients used Message Passing Interface (MPI) protocols on the localhost, as at the time the Gromacs cores were not designed to be used with multiple threads.[119] This made Folding@home the first distributed computing project to utilize MPI, as it had previously been reserved only for supercomputers.[123] The MPI-based clients worked well in Unix-based operating systems such as Linux and Mac's OS-X, but was particularly troublesome in Windows.[153][123] Despite these difficulties, SMP1 generated significant results that would have been impossible otherwise and which represented a landmark in the simulation of protein folding.[153]
The second generation of the SMP client was released as an open beta on January 24, 2010, and subsequently replaced SMP1,[154] although its interface was remained based on the command-line. The SMP2 client exchanges the complex MPI for threads, which removes much of the overhead of keeping the cores synchronized.[154][155] The SMP2 client also supports a bonus points system, which non-linearly rewards additional points to donors for quick WU returns and for contributing to next-generation capabilities.[119] Donors who run the SMP2 client receive these extra points if they use a passkey and maintained an 80% successful return of Work Units.[154]
SMP2 also supports extra-large Work Units for users with powerful sixteen-core CPUs or better.[156] While these WUs consume even more RAM and have more network usage than regular SMP WUs, users who run these are rewarded with a 20% increase over SMP2's bonus point system.[157][158] These powerful computers allow for simulations to be performed on Folding@home that had previously required the use of supercomputing clusters. There is a great scientific need to run these simulations out to long timescales as quickly as possible, so the additional bonus points also serves as an incentive for rapid completions of Work Units. This allows the Pande lab to perform studies of larger molecular systems that would not have been possible anywhere else on Folding@home.[158]
V7

The v7 client is the seventh and latest generation of the Folding@home software, currently under development, but available for open beta testing.[4] V7 is a complete rewrite and unification of the previous clients for Microsoft Windows, Mac OS X and Linux operating systems.[159][160] Following its predecessors, v7 runs Folding@home in the background at very low priority, which allows other applications to use CPU resources as they need.[159] The v7 client is designed to make the installation, start-up, and operation user-friendly for novices, as well as offer greater scientific flexibility than previous clients.[159][161] The Pande lab's goal was to make v7 the recommended client by January 2012, but as of February 2012, the v7 client remains in open beta testing.[162]
V7 consists of several elements. The user interacts with v7's GUI, known as FAHControl. It has Novice, Advanced, and Expert user interface modes, and has the ability to monitor, configure, and control many remote folding clients from a single computer.[159] FAHControl can monitor and direct FAHClient, which runs behind the scenes and in turn manages each FAHSlot (or "slot"). These slots act as replacements for the previously distinct FAH clients, as they may be of Uniprocessor, SMP, or GPU type. Each slot also contains a core and data associated with it, and can download, process, and upload Work Units independently.[159][163] The FAHViewer function, modeled after the PS3 viewer, displays a real-time 3D rendering, if available, of the protein currently being processed.[159][163]
Work Units
The Work Unit (WU) is the protein data that the client is being asked to process. Work Units are a fraction of the simulation calculating the rate of transitions between the states in a Markov state model. The client connects to the Folding@home server to retrieve this Work Unit and may also download an appropriate core. Once completed, the results are returned and the respective credit points are awarded, and this cycle then repeats automatically.[80] During transfer, all Work Units are validated through the use of 2048-bit digital signatures.[129] These WUs have associated deadlines and credit (point) value. If this deadline is exceeded, the user may not get credit and the unit will be reissued to someone else.[129] As protein folding is serial in nature and each WU is generated from its predecessor, this allows the overall simulation process to proceed normally if a WU is not returned after a certain period of time.[129] Due to these deadlines, the minimum system requirements for Folding@home is a Pentium 3 450 MHz CPU with SSE or newer.[129] However, Work Units for high performance clients have a much shorter deadline than those for the uniprocessor client, as a major part of the scientific benefit is dependent on rapidly completing simulations.[119]
Before public release, Work Units go through several quality assurance steps to keep problematic WUs from becoming fully available.[164] But unlike particular BOINC projects such as SETI@home, Folding@home's Work Units are normally processed only once, except in the rare event that errors occur during processing of a WU. If this occurs for three different donors,[165] it is automatically pulled from distribution.[166][167] Topics in the Folding@home forum can be used to differentiate between problematic hardware and an actual bad Work Unit.[168]
Cores
Specialized scientific computer programs, referred to as "cores," perform the calculations on the Work Unit behind the scenes.[80] Folding@home's cores modified and optimized versions of molecular dynamics programs, including GROMACS, AMBER, TINKER, CPMD, SHARPEN, ProtoMol, BrookGPU and Desmond.[169][170] Some of these cores perform explicit atom-by-atom molecular dynamics calculations,[171] while others perform implicit solvation methods, which treat atoms as a mathematical continuum.[137][172] These cores are open-source software or are under similar licenses,[173][174] and are verified during download by 2048-bit digital signatures. While the same core can be used by various versions of the client, separating the core from the client enables the scientific methods to be updated automatically as needed without a client update.[80]
Comparison to other molecular systems
Rosetta@home is a distributed computing project aimed at protein structure prediction and is one of the most successful approaches to this problem.[175] Folding@home and Rosetta@home address very different molecular questions.[176] Although Rosetta@home does not provide information into how proteins fold, it does predict the protein's most likely final structure, which in some cases is used as a basis for Folding@home's projects.[177][178] Rosetta's predictions can help FAH simulate the folding of larger proteins more efficiently. Folding@home can also verify Rosetta@home's results and find additional atomistic details of the protein's kinetics and folding pathway,[177] which is intrinsically much more difficult.[25] Folding@home's accurate simulations have also suggesting important novel implications into the fields of protein folding, structure prediction, and certain folding experiments,[179] and have shown that Rosetta's structure prediction may benefit from thermodynamic sampling aspects of protein folding mechanisms.[180][181]
Folding@home also compares well to Anton, a powerful supercomputer which uses specialized hardware to produce a small number of ultra-long molecular trajectories. It is unique in this ability, and like Folding@home, has also improved particular long-held theories of protein folding. Its longer simulations, while computationally expensive, contain more phase space than any one of Folding@home's many shorter trajectories, which allows Anton to perform a thorough exploration of the required space.[182] As of October 2011, Anton and FAH are the two most powerful molecular dynamics systems,[183] and Anton has also run simulations out to the millisecond range.[184][185] In 2011, the Pande lab built a Markov state model from one of Anton's simulations.[182] It demonstrated that there was little difference between MSMs built from Anton's fewer long trajectories and one assembled from Folding@home's many shorter trajectories. Their analysis also showed that Folding@home's Markov state models significantly improve the analysis of these longer simulations, such as revealing additional relevant folding pathways and information into how the protein carries out its biological function.[182] Folding@home is running further analysis on one of Anton's simulations to better determine how its approaches compare to Anton's methods.[186] It is probable that a combination of Anton's and FAH's simulation methods would be very beneficial,[182] and Pande looks forward to see how Anton and FAH can be used together.[183]
See also
- Storage@home
- List of distributed computing projects
- Software for molecular modeling
- Molecular modeling on GPUs
- Rosetta@home
- Blue Gene
- Molecular dynamics
- Computational biology
External links
- Folding@home homepage
- Folding@home official blog
- Folding@home forum
- Folding@home statistics
- List of publications from Folding@home's results
- "Futures In Biotech" episode (for newcomers)
- 2009 Interview with Vijay Pande about Folding@home Project
- Video of record-breaking 1.5ms protein fold
- Pande lab's OpenMM molecular dynamics library
- Simple multimedia presentation about Folding@home
- Folding@home Wiki
- Wikipedia Team
- Folding@home -Facebook
Notes
Note 1: It should be noted that accurate measurement of the speed of a supercomputer does not necessarily equate to scientific productivity. Supercomputers are typically tested for brief periods using the legacy LINPACK benchmark. This brief CPU testing is not an accurate indication of their prolonged performance over real-world tasks.[108][187]
References
- ^ a b c d e Pande lab (2011-09-19). "Download the Folding@home Software Application". Stanford University. Retrieved 2011-08-31.
- ^ a b Pande lab. "High Performance Clients". Stanford University. Retrieved 2011-08-31.
- ^ "Folding@home for PlayStation3". Sony. 2008. Retrieved 2011-08-31.
- ^ a b "Folding@Home v7 Client Beta Release Page". Stanford University. Retrieved 2011-09-19.
- ^ Joseph Coffland (2012-02-15). "FAHClient V7.1.48 released (6th Open-Beta)". Retrieved 2012-02-15.
- ^ a b c Pande lab. "Folding@home Distributed Computing Client". Stanford University. Retrieved 2010-08-26.
- ^ Imran "ihaque" Haque (Pande lab member) (2010-08-11). "Re: FAH really doing anything?". Retrieved 2011-08-23.
- ^ a b Bojan Zagrovic, Christopher D. Snow, Michael R. Shirts, and Vijay S. Pande. (2002). "Simulation of Folding of a Small Alpha-helical Protein in Atomistic Detail using Worldwide distributed Computing". Journal of Molecular Biology. 323 (5): 927–937. doi:10.1016/S0022-2836(02)00997-X. PMID 12417204.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Pande lab. "Folding@Home Executive summary" (PDF). Stanford University. Retrieved 2011-10-04.
- ^ Pande lab (2011). "Folding@home - Main". Stanford University. Retrieved 2011-10-04.
- ^ Antonella De Jaco, Michael Z. Lin, Noga Dubi, Davide Comoletti, Meghan T. Miller, Shelley Camp, Mark Ellisman, Margaret T. Butko, Roger Y. Tsien, and Palmer Taylor (2010). "Neuroligin Trafficking Deficiencies Arising from Mutations in the a/ß-Hydrolase Fold Protein Family". Journal of Biological Chemistry. 285 (37): 28674–28682. doi:10.1074/jbc.M110.139519. PMC 2937894. PMID 20615874.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c d e f g h i j k l m Pande lab. "Folding@home Diseases Studied FAQ" (FAQ). Stanford University. Retrieved 2011-09-23.
- ^ Bishnu Mukhopadhyay and Hridoy Bairagya (2011). Journal of Biomolecular Structure & Dynamics. 28 (4). ISSN 0739-1102 http://www.jbsdonline.com/mc_images/category/4308/29-bishnu_jbsd_28_4.pdf.
{{cite journal}}: Missing or empty|title=(help) - ^ a b Pande lab (2011-08-05). "Folding@home - Papers". Stanford University. Retrieved 2011-10-09.
- ^ a b c V. S. Pande, K. Beauchamp, and G. R. Bowman (2010). "Everything you wanted to know about Markov State Models but were afraid to ask". Methods. 52 (1): 99–105. doi:10.1016/j.ymeth.2010.06.002. PMC 2933958. PMID 20570730.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pande lab (2011). "Folding@home - About". Stanford University. Retrieved 2011-08-31.
- ^ F. Chiti, N. Taddei, G. Ramponi, and C. M. Dobson (2003). "Rationalization of the effects of mutations on peptide and protein aggregation rates". Nature: 805–808. PMID 12917692.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f g G. Bowman, V. Volez, and V. S. Pande (2011). "Taming the complexity of protein folding". Current Opinion in Structural Biology. 21 (1): 4–11. doi:10.1016/j.sbi.2010.10.006. PMC 3042729. PMID 21081274.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Leila M Luheshi, Damian Crowther, Christopher Dobson (2008). "Protein misfolding and disease: from the test tube to the organism". Current Opinion in Chemical Biology. 12 (1): 25–31. doi:10.1016/j.cbpa.2008.02.011. PMID 18295611.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Yiwen Chen, Feng Ding, Huifen Nie, Adrian W. Serohijos, Shantanu Sharma, Kyle C. Wilcox, Shuangye Yin, Nikolay V. Dokholyan (2008). "Protein folding: Then and now". Archives of Biochemistry and Biophysics. 469 (1): 4–19. doi:10.1016/j.abb.2007.05.014.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fabrizio Marinelli1,2, Fabio Pietrucci1, Alessandro Laio1*, Stefano Piana (2009). "A Kinetic Model of Trp-Cage Folding from Multiple Biased Molecular Dynamics Simulations". PLoS Computational Biology. 8 (5). doi:10.1371/journal.pcbi.1000452.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Joseph Bordogna (2001). "The 21st Century Engineer". IEEE Spectrum. 38 (1): 17.
- ^ "Cutting through the Protein Knots". Astrobiology Magazine. 2002-10-24. Retrieved 2012-02-22.
- ^ a b c d e f Vijay S. Pande, Ian Baker, Jarrod Chapman, Sidney P. Elmer, Siraj Khaliq, Stefan M. Larson, Young Min Rhee, Michael R. Shirts, Christopher D. Snow, Eric J. Sorin, Bojan Zagrovic (2002). "Atomistic protein folding simulations on the submillisecond timescale using worldwide distributed computing". Biopolymers. 68 (1): 91–109. doi:10.1002/bip.10219. PMID 12579582.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b C. D. Snow, E. J. Sorin, Y. M. Rhee, and V. S. Pande. (2005). "How well can simulation predict protein folding kinetics and thermodynamics?". Annual Reviews of Biophysics. 34: 43–69. doi:10.1146/annurev.biophys.34.040204.144447. PMID 15869383.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vincent A. Voelz, Gregory R. Bowman, Kyle Beauchamp and Vijay S. Pande (2010). "Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1–39)". Journal of the American Chemical Society. 132 (5): 1526–1528. doi:10.1021/ja9090353. PMC 2835335. PMID 20070076.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Greg Bowman awarded the 2010 Kuhn Paradigm Shift Award". SimTK: MSMBuilder. 2010-03-29. Retrieved 2011-11-30.
- ^ "Thomas Kuhn Paradigm Shift Award". ACS: Computers in Chemistry. Retrieved 2011-11-30.
- ^ a b c d e f g Pande lab. "Folding@Home Press FAQ" (FAQ). Stanford University. Retrieved 2011-08-31.
- ^ a b Pande lab. "Project 3430 Description". Stanford University. Retrieved 2011-11-30.
- ^ Christian "schwancr" Schwantes (Pande lab member) (2011-08-15). "Projects 7808 and 7809 to full fah". Retrieved 2011-10-16.
- ^ Del Lucent, V. Vishal, and Vijay S. Pande (2007). "Protein folding under confinement: A role for solvent". Proceedings of the National Academy of Sciences of the United States of America. 104 (25): 10430–10434. doi:10.1073/pnas.0608256104.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Eric Sorin and Vijay Pande (2006). "Nanotube Confinement Denatures Protein Helices" (PDF). Journal of the American Chemical Society. 128 (19): 6316–6317. doi:10.1021/ja060917j.
- ^ Pande lab. "Project 2974 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 6811 Description". Stanford University. Retrieved 2011-09-27.
- ^ G. R. Bowman and V. S. Pande (2010). "Protein folded states are kinetic hubs". Proceedings of the National Academy of Sciences, USA. 107 (24): 10890–10895. Bibcode:2010PNAS..10710890B. doi:10.1073/pnas.1003962107. PMC 2890711. PMID 20534497.
- ^ V. S. Pande (2010). "A simple theory of protein folding kinetics". Physical Review Letters. 105 (19). Bibcode:2010PhRvL.105s8101P. doi:10.1103/PhysRevLett.105.198101.
- ^ Christopher D. Snow, Houbi Ngyen, Vijay S. Pande, and Martin Gruebele (2002). "Absolute comparison of simulated and experimental protein-folding dynamics" (PDF). Nature. 420 (6911): 102–106. doi:10.1038/nature01160. PMID 12422224.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Vijay Pande (2008-04-23). "Folding@home and Simbios". Retrieved 2011-11-09.
- ^ a b Vijay Pande (2011-10-25). "Re: Suggested Changes to F@h Website". Retrieved 2011-10-25.
- ^ Sarah Jane Keller (2011-11-18). "New Stanford software takes Folding@home's biological research to supercomputers". Stanford University. Retrieved 2011-11-30.
- ^ Pascale Lacor, Maria Buniel, Paul Furlow, Antonio Clemente, Pauline Velasco, Margaret Wood, Kirsten Viola, and William Klein (2007). "Aß Oligomer-Induced Aberrations in Synapse Composition, Shape, and Density Provide a Molecular Basis for Loss of Connectivity in Alzheimer's Disease". Journal of Neuroscience. 27 (4): 796–807. doi:10.1523/JNEUROSCI.3501-06.2007. PMID 17251419.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ P. Novick, J. Rajadas, C.W. Liu, N. W. Kelley, M. Inayathullah, and V. S. Pande (2011). Buehler, Markus J. (ed.). "Rationally Designed Turn Promoting Mutation in the Amyloid-ß Peptide Sequence Stabilizes Oligomers in Solution". PLoS ONE. 6 (7): e21776. doi:10.1371/journal.pone.0021776. PMC 3142112. PMID 21799748.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c d Nicholas W. Kelley, V. Vishal, Grant A. Krafft, and Vijay S. Pande. (2008). "Simulating oligomerization at experimental concentrations and long timescales: A Markov state model approach". Journal of Chemical Physics. 129 (21): 214707. Bibcode:2008JChPh.129u4707K. doi:10.1063/1.3010881. PMC 2674793. PMID 19063575.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Vijay Pande (2011-08-05). "Results page updated – new key result published in our work in Alzheimer's Disease". Retrieved 2011-09-10.
- ^ Pande lab. "Project 6802 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 6871 Description". Stanford University. Retrieved 2011-09-27.
- ^ Vijay Pande (2008-12-18). "New FAH results on possible new Alzheimer's drug presented". Retrieved 2011-09-23.
- ^ Pande lab. "Project 700 Description". Stanford University. Retrieved 2011-09-27.
- ^ Walker FO (2007). "Huntington's disease". Lancet. 369 (9557): 220. doi:10.1016/S0140-6736(07)60111-1. PMID 17240289.
- ^ a b Nicholas W. Kelley, Xuhui Huang, Stephen Tam, Christoph Spiess, Judith Frydman and Vijay S. Pande (2009). "The predicted structure of the headpiece of the Huntingtin protein and its implications on Huntingtin aggregation". Journal of Molecular Biology. 388 (5): 919–27. doi:10.1016/j.jmb.2009.01.032. PMC 2677131. PMID 19361448.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Orr HT (2004). "Neurodegenerative disease: neuron protection agency" (PDF). Nature. 431 (7010): 747–8. doi:10.1038/431747a. PMID 15483586.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ M Hollstein, D Sidransky, B Vogelstein and CC Harris (1991). "p53 mutations in human cancers". Science. 253 (5015): 49–53. doi:10.1126/science.1905840. PMID 1905840.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ L.T. Chong, W. C. Swope, J. W. Pitera, and V. S. Pande (2006). "A novel approach for computational alanine scanning: application to the p53 oligomerization domain". Journal of Molecular Biology. 357 (3): 1039–1049. doi:10.1016/j.jmb.2005.12.083. PMID 16457841.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Vijay Pande (2005-01-16). "First results from Folding@Home cancer project published". Wayback Machine. Archived from the original on 2007-07-06. Retrieved 2011-11-09.
- ^ L. T. Chong, C. D. Snow, Y. M. Rhee, and V. S. Pande. (2004). "Dimerization of the p53 Oligomerization Domain: Identification of a Folding Nucleus by Molecular Dynamics Simulations". Journal of Molecular Biology. 345 (4): 869–878. doi:10.1016/j.jmb.2004.10.083. PMID 15588832.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Vijay Pande (2007-09-28). "Nanomedicine center". Retrieved 2011-09-23.
- ^ Vijay Pande (2009-12-22). "Release of new Protomol (Core B4) WUs". Retrieved 2011-09-23.
- ^ Pande lab. "Project 180 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 7600 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 10113 Description". Stanford University. Retrieved 2011-09-27.
- ^ Sanghyun Park, Randall J. Radmer, Teri E. Klein, and Vijay S. Pande (2005). "A New Set of Molecular Mechanics Parameters for Hydroxyproline and Its Use in Molecular Dynamics Simulations of Collagen-Like Peptides". Journal of Computational Chemistry. 26 (15): 1612–1616. doi:10.1002/jcc.20301. PMID 16170799.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Peter Kasson. "Membrane Fusion". Stanford University. Retrieved 2011-12-02.
- ^ Gregory Bowman (Pande Group Member). "Project 10125". Stanford University. Retrieved 2011-12-02.
- ^ a b "Home computers aid disease fight". BBC News. 2004-09-18. Retrieved 2011-12-01.
- ^ a b Guha Jayachandran, M. R. Shirts, S. Park, and V. S. Pande (2006). "Parallelized-Over-Parts Computation of Absolute Binding Free Energy with Docking and Molecular Dynamics". Journal of Chemical Physics. 125 (8): 084901. Bibcode:2006JChPh.125h4901J. doi:10.1063/1.2221680. PMID 16965051.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Hideaki Fujutani, Yoshiaki Tanida, Masakatsu Ito, Guha Jayachandran, Christopher D. Snow, Michael R. Shirts, Eric J. Sorin, and Vijay S. Pande (2005). "Direct calculation of the binding free energies of FKBP ligands using the Fujitsu BioServer massively parallel computer". Journal of Chemical Physics. 123 (8): 084108. Bibcode:2005JChPh.123h4108F. doi:10.1063/1.1999637. PMID 16164283.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Michael Shirts (Pande Group Member) (2011-05-23). "New projects 3865-3867 in beta". Retrieved 2011-12-01.
- ^ Pande lab. "Project 3855 Description". Stanford University. Retrieved 2011-12-01.
- ^ Pande lab. "Project 2450 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 10115 Description". Stanford University. Retrieved 2011-09-27.
- ^ Pande lab. "Project 5765 Description". Stanford University. Retrieved 2011-12-02.
- ^ Vijay Pande (2007-10-21). "Fun fact: FAH growth over time". Retrieved 2011-10-21.
- ^ Pande lab. "Active CPUs" (Image). Stanford University. Retrieved 2011-08-30.
- ^ a b c d Pande lab (updated daily). "Client Statistics by OS". Stanford University. Retrieved 2011-12-23.
{{cite web}}: Check date values in:|date=(help) - ^ "BOINC Combined Credit Overview". Retrieved 2012-01-26.
- ^ Michael R. Shirts and Vijay S. Pande (2001). "Mathematical Foundations of Coupled Parallel Simulations". Physical Review Letters. 86 (22): 4983–4987. Bibcode:2001PhRvL..86.4983S. doi:10.1103/PhysRevLett.86.4983. PMID 11384401.
- ^ Joshua Topolsky (2007-10-31). "Folding@Home recognized by Guinness World Records". Retrieved 2007-11-05.
- ^ Caroline Hadley (2004). "Biologists think bigger". EMBO reports. 12 (5): 236–238. doi:10.1038/sj.embor.7400108.
- ^ a b c d e f Adam Beberg, Daniel Ensign, Guha Jayachandran, Siraj Khaliq, Vijay Pande (2009). "Folding@home: Lessons From Eight Years of Volunteer Distributed Computing" (PDF). Parallel & Distributed Processing, IEEE International Symposium: 1–8. doi:10.1109/IPDPS.2009.5160922. ISSN 1530-2075.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shankland, Stephen (March 22, 2002). "Google takes on supercomputing". CNet News.
- ^ a b c d "Futures in Biotech 27: Folding@home at 1.3 Petaflops" (Interview, webcast). 2007-12-28.
- ^ ChelseaOilman (2005-12-30). "Google is after your CPU cycles". Retrieved 2011-09-06.
- ^ Google (2007). "Your computer's idle time is too precious to waste". Retrieved 2011-09-06.
{{cite news}}:|author=has generic name (help) - ^ "Genome@home Updates". 2002-03-04. Retrieved 2011-09-05.
- ^ Pande lab. "Genome@home FAQ" (FAQ). Stanford University. Retrieved 2011-09-05.
- ^ "TOP500 List - June 2007". Top500. 06-2007. Retrieved 2011-11-11.
{{cite web}}: Check date values in:|date=(help) - ^ "TOP500 List - November 2007". Top500. 11-2007. Retrieved 2011-11-11.
{{cite web}}: Check date values in:|date=(help) - ^ a b "TOP500 List - June 2008". Top500. 06-2008. Retrieved 2011-11-11.
{{cite web}}: Check date values in:|date=(help) - ^ "TOP500 List - November 2008". Top500. 11-2008. Retrieved 2011-11-11.
{{cite web}}: Check date values in:|date=(help) - ^ "TOP500 List - June 2011". Top500. 06-2011. Retrieved 2011-11-11.
{{cite web}}: Check date values in:|date=(help) - ^ Vijay Pande (2007-09-16). "Crossing the petaFLOPS barrier". Retrieved 2011-08-28.
- ^ David Nagel (2007-09-19). "Folding@home Achieves Petaflop Milestone - PS3 owners help scientists speed up their research". Retrieved 2011-09-06.
- ^ Mark Wilson (2007-03-25). "PS3 Folding@Home TFLOP Rating Demoted by 50%, PFLOPS Still Possible". Retrieved 2011-09-14.
- ^ Tim Hanlon (2007-03-09). "Playstation 3 continues to top Folding@Home statistics". Retrieved 2011-09-14.
- ^ "Folding@Home reach 2 Petaflops". 2008-05-08. Retrieved 2011-09-23.
- ^ "NVIDIA Achieves Monumental Folding@Home Milestone With Cuda". 2008-08-26. Retrieved 2011-09-06.
- ^ "3 PetaFLOP barrier". 2008-08-19. Retrieved 2011-09-23.
- ^ Vijay Pande (2009-02-18). "Folding@home Passes the 5 petaFLOP Mark". Retrieved 2011-08-31.
- ^ "Crossing the 5 petaFLOPS barrier". 2009-02-18. Retrieved 2011-09-23.
- ^ Jesse_V (2011-11-10). "Six Native PetaFLOPS". Retrieved 2011-11-11.
- ^ Vijay Pande (2009-03-18). "FLOPS". Retrieved 2011-10-11.
- ^ a b Pande lab (2009-04-04). "Folding@home FLOP FAQ" (FAQ). Stanford University. Retrieved 2011-08-28.
- ^ a b c d e Pande lab (2009-02-05). "PS3 FAQ" (FAQ). Stanford University. Retrieved 2011-09-05.
- ^ a b Pande lab (2011-02-16). "Folding@home Points FAQ" (FAQ). Stanford University. Retrieved 2011-08-31.
- ^ a b Pande lab (2011-02-16). "Folding@home Points FAQ (New Benchmark Machine -- January 2010)" (FAQ). Stanford University. Retrieved 2011-08-31.
- ^ Peter Kasson (Pande lab member) (2010-01-24). "Upcoming Release of SMP2 Cores". Retrieved 2011-08-31.
- ^ a b Bruce Borden (bruce) (2010-04-07). "Re: Answers to: Reasons for not using F@H". Retrieved 2011-09-05.
- ^ Pande lab (2011-05-24). "Folding@home Passkey FAQ" (FAQ). Stanford University. Retrieved 2011-09-06.
- ^ Tim "7im" Braun (2011-09-29). "Re: Passkey when changing username". Retrieved 2011-10-02.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ "Default Team". Retrieved 2011-09-06.
- ^ MtM (2009-12-17). "Re: New to F@H need startup info". Retrieved 2011-09-30.
- ^ a b "Re: Why join a team?". 2009-08-28. Retrieved 2011-09-08.
- ^ "Official Extreme Overclocking Folding@Home Team Forum". Extreme Overclocking. Retrieved 2011-09-08.
- ^ Norman Chan (2009-04-06). "Help Maximum PC's Folding Team Win the Next Chimp Challenge!". Retrieved 2011-09-06.
- ^ "Team 24 Folding at Home - March Challenge". 2011-02-20. Retrieved 2011-09-06.
- ^ "Announcing the F@H Prize". Immortality Institute. Retrieved 2011-09-06.
- ^ "Third Party Contributions - Stats Pages". 2011. Retrieved 2011-08-07.
- ^ a b c d e f g Pande lab (2010-12-11). "Folding@home SMP FAQ" (FAQ). Stanford University. Retrieved 2011-08-31.
- ^ a b Vijay Pande (2008-05-23). "GPU news (about GPU1, GPU2, & NVIDIA support)". Retrieved 2011-09-08.
- ^ Travis Desell1, Anthony Waters, Malik Magdon-Ismail, Boleslaw K. Szymanski, Carlos A. Varela, Matthew Newby, Heidi Newberg, Andreas Przystawik, and David Anderson (2009). "Accelerating the MilkyWay@Home volunteer computing project with GPUs". 8th International Conference on Parallel Processing and Applied Mathematics (PPAM 2009). doi:10.1.1.158.7614.
{{cite journal}}: Check|doi=value (help)CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Lou Kesten, Associated Press (2007-03-26). "Week in video-game news: 'God of War II' storms the PS2; a PS3 research project". Pittsburgh Post-Gazette. Retrieved 2011-12-13.
- ^ a b c Vijay Pande (2008-03-08). "New Windows client/core development (SMP and classic clients)". Retrieved 2011-09-30.
- ^ a b c d Pande lab (2011-02-10). "Windows Uniprocessor Client Installation Guide". Stanford University. Retrieved 2011-09-05.
- ^ a b John Naylor (2008-02-09). "Answers to: Reasons for not using F@H". Retrieved 2011-09-05.
- ^ Bruce Borden (bruce) (2011-09-11). "Re: F@h Advertisement Techniques". Retrieved 2011-10-18.
- ^ Tim "7im" Braun (2008-09-28). "Answers to: Reasons for not using F@H". Retrieved 2011-09-05.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ Bruce Borden (bruce) (2008-07-28). "Re: Answers to: Reasons for not using F@H". Retrieved 2011-09-05.
- ^ a b c d e f g Pande lab (2011-08-18). "Folding@home Main FAQ" (FAQ). Stanford University. Retrieved 2011-10-23.
- ^ Vijay Pande (2009-08-20). "Importance of software and data integrity". Retrieved 2011-10-19.
- ^ a b c Bruce Borden (bruce) (2011-07-18). "Re: Advice for a new user". Retrieved 2011-09-11.
- ^ a b Pande lab (2009-11-19). "Uninstalling Folding@home FAQ". Stanford University. Retrieved 2011-09-21.
- ^ M. R. Shirts and V. S. Pande. (2000). "Screen Savers of the World, Unite!". Science. 290 (5498): 1903–1904. doi:10.1126/science.290.5498.1903. PMID 17742054.
- ^ a b c d e Pande lab (2010-05-13). "High Performance FAQ" (FAQ). Stanford University. Retrieved 2011-09-05.
- ^ John D. Owens, David Luebke, Naga Govindaraju, Mark Harris, Jens Krüger, Aaron Lefohn, Timothy J. Purcell (2007). "A Survey of General-Purpose Computation on Graphics Hardware". Computer Graphics Forum. 26 (1): 80–113.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ P. Eastman and V. S. Pande (2010). "OpenMM: A Hardware Abstraction Layer for Molecular Simulations". Computing in Science & Engineering. 12 (4): 34–39. doi:10.1109/MCSE.2010.27.
- ^ a b M. S. Friedrichs, P. Eastman, V. Vaidyanathan, M. Houston, S. LeGrand, A. L. Beberg, D. L. Ensign, C. M. Bruns, V. S. Pande (2009). "Accelerating Molecular Dynamic Simulation on Graphics Processing Units". Journal of Computational Chemistry. 30 (6): 864–72. doi:10.1002/jcc.21209. PMC 2724265. PMID 19191337.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ I. Haque and V. S. Pande (2010). "Hard Data on Soft Errors: A Large-Scale Assessment of Real-World Error Rates in GPGPU". 2010 10th IEEE/ACM International Conference on Cluster, Cloud and Grid Computing (CCGrid): 691–696. doi:10.1109/CCGRID.2010.84.
- ^ a b Pande lab (2011-03-18). "ATI FAQ" (FAQ). Stanford University. Retrieved 2011-08-31.
- ^ Vijay Pande (2008-05-27). "More info about the GPU1 to GPU2 transition". Retrieved 2011-09-07.
- ^ Vijay Pande (2008-04-10). "GPU2 open beta". Retrieved 2011-09-07.
- ^ Vijay Pande (2008-06-06). "GPU1 has been retired, GPU2 for NVIDIA release nearing". Retrieved 2011-09-07.
- ^ Vijay Pande (2008-04-15). "Updates to the Download page/GPU2 goes live". Retrieved 2011-09-07.
- ^ Vijay Pande (2008-04-11). "GPU2 open beta going well". Retrieved 2011-09-07.
- ^ Vijay Pande (2008-01-25). "Code Development Updates". Retrieved 2011-09-07.
- ^ a b Vijay Pande (2010-04-24). "Prepping for the GPU3 rolling: new client and NVIDIA FAH GPU clients will (in the future) need CUDA 2.2 or later". Retrieved 2011-09-08.
- ^ Vijay Pande (2010-05-25). "Folding@home: Open beta release of the GPU3 client/core". Retrieved 2011-09-07.
- ^ Joseph Coffland (2011-10-13). "Re: FAHClient V7.1.38 released (4th Open-Beta)". Retrieved 2011-10-15.
- ^ "NVIDIA GPU3 Linux/Wine Headless Install Guide". 2008-11-08. Retrieved 2011-09-05.
- ^ a b David E. Williams (2006-10-20). "PlayStation's serious side: Fighting disease". CNN. Retrieved 2011-10-16.
- ^ a b Edgar Luttmann, Daniel L. Ensign, Vishal Vaidyanathan, Mike Houston, Noam Rimon, Jeppe Øland, Guha Jayachandran, Mark Friedrichs, Vijay S. Pande (2008). "Accelerating Molecular Dynamic Simulation on the Cell processor and PlayStation 3". Journal of Computational Chemistry. 30 (2): 268–274. doi:10.1002/jcc.21054. PMID 18615421.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ozzfan (2007-10-01). "Can I have multiple CPUs work on the same task (workunit)?". BOINC. Retrieved 2011-10-09.
- ^ a b c d Vijay Pande (2008-06-15). "What does the SMP core do?". Retrieved 2011-09-07.
- ^ a b c Peter Kasson (Pande lab member) (2010-01-24). "upcoming release of SMP2 cores". Retrieved 2011-09-30.
- ^ Vijay Pande (2009-06-17). "How does FAH code development and sysadmin get done?". Retrieved 2011-10-14.
- ^ Vijay Pande (2011-02-07). "Update on "bigadv-16", the new bigadv rollout". Retrieved 2012-02-09.
- ^ Vijay Pande (2011-07-02). "Change in the points system for bigadv work units". Retrieved 2011-10-09.
- ^ a b Peter Kasson (Pande lab member) (2009-07-15). "new release: extra-large work units". Retrieved 2011-10-09.
- ^ a b c d e f Vijay Pande (2011-03-29). "Client version 7 now in open beta". Retrieved 2011-08-14.
- ^ Pande lab (2011). "Client FAQ" (FAQ). Stanford University. Retrieved 2011-08-14.
- ^ Vijay Pande (2011-03-31). "Core 16 for ATI released; also note on NVIDIA GPU support for older boards". Retrieved 2011-09-07.
- ^ Joseph Coffland (2011-09-19). "FAHClient V7.1.33 released (3rd Open-Beta)". Retrieved 2011-09-19.
- ^ a b Pande lab (2011-09-23). "Windows (FAH V7) Install Guide". Stanford University. Retrieved 2011-10-09.
- ^ Vijay Pande (2011-04-05). "More transparency in testing". Retrieved 2011-10-14.
- ^ Bruce Borden (bruce) (2011-08-07). "Re: Gromacs Cannot Continue Further". Retrieved 2011-08-07.
- ^ Bruce Borden (bruce) (2011-08-09). "Re: Project: 6053 (Run 1, Clone 194, Gen 357)". Retrieved 2011-08-09.
- ^ PantherX (2011-10-01). "Re: Project 6803: (Run 4, Clone 66, Gen 255)". Retrieved 2011-10-09.
- ^ PantherX (2010-10-31). "Troubleshooting Bad WUs". Retrieved 2011-08-07.
- ^ "Cores - FaHWiki" (FAQ). Retrieved 2007-11-06.
- ^ Pande lab (2005-10-16). "Folding@home with QMD core FAQ" (FAQ). Stanford University. Retrieved 2006-12-03.
- ^ Vijay Pande (2007-09-26). "How FAH works: Molecular dynamics". Retrieved 2011-10-14.
- ^ Roux B, Simonson T (1999). "Implicit solvent models". Biophys. Chem. 78 (1–2): 1–20. doi:10.1016/S0301-4622(98)00226-9. ISSN 0301-4622. PMID 17030302.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Pande lab (2010-02-03). "Folding@home Open Source FAQ" (FAQ). Stanford University. Retrieved 2011-09-11.
- ^ Vijay Pande (2011-09-06). "Re: Utilizing this resource". Retrieved 2011-09-11.
- ^ Gregory R. Bowman and Vijay S. Pande (2009). "Simulated tempering yields insight into the low-resolution Rosetta scoring function". Proteins: Structure, Function, and Bioinformatics. 74 (3): 777–88. doi:10.1002/prot.22210. PMID 18767152.
- ^ Vijay Pande (2006-03-05). "Rosetta and Folding". Retrieved 2011-11-09.
- ^ a b TJ Lane (Pande lab member) (2011-06-09). "Re: Course grained Protein folding in under 10 minutes". Retrieved 2011-10-15.
- ^ Bruce Borden (bruce) (2011-09-24). "Re: New Invention Unravels Mystery of Protein Folding". Retrieved 2011-10-29.
- ^ Bojan Zagrovic, Christopher D. Snow, Siraj Khaliq, Michael R. Shirts, and Vijay S. Pande (2002). "Native-like Mean Structure in the Unfolded Ensemble of Small Proteins". Journal of Molecular Biology. 323 (1): 153–164. doi:10.1016/S0022-2836(02)00888-4. PMID 12368107.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ G. R. Bowman and V. S. Pande (2009). Hofmann, Andreas (ed.). "The Roles of Entropy and Kinetics in Structure Prediction". PLoS ONE. 4 (6): e5840. Bibcode:2009PLoSO...4.5840B. doi:10.1371/journal.pone.0005840. PMC 2688754. PMID 19513117.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Vijay Pande (2008-04-26). "Re: collaborating with competition". Retrieved 2011-11-09.
- ^ a b c d Thomas J. Lane, Gregory R. Bowman, Kyle A Beauchamp, Vincent Alvin Voelz, and Vijay S. Pande (2011). "Markov State Model Reveals Folding and Functional Dynamics in Ultra-Long MD Trajectories". Journal of the American Chemical Society. doi:10.1021/ja207470h.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Vijay Pande (2011-10-13). "Comparison between FAH and Anton's approaches". Retrieved 2011-10-13.
- ^ Heidi Ledford (2010-10-14). "Supercomputer sets protein-folding record". Nature News. doi:10.1038/news.2010.541. Retrieved 2011-10-15.
- ^ David E. Shaw, Paul Maragakis, Kresten Lindorff-Larsen, Stefano Piana, Ron O. Dror, Michael P. Eastwood, Joseph A. Bank, John M. Jumper, John K. Salmon, Yibing Shan, and Willy Wriggers (2010). "Atomic-Level Characterization of the Structural Dynamics of Proteins". Science. 330 (6002): 341–346. doi:10.1126/science.1187409. PMID 20947758.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pande lab (2011). "Project 7610 Description". Retrieved 2011-10-14.
- ^ Vijay Pande (2008-11-09). "Re: ATI and NVIDIA stats vs. PPD numbers". Retrieved 2011-09-22.
- Distributed computing projects
- Molecular dynamics software
- Molecular modelling software
- Molecular modelling
- Simulation software
- Protein folds
- Protein structure
- Computational biology
- Mathematical and theoretical biology
- Bioinformatics
- Computational chemistry
- Hidden Markov models
- Data mining and machine learning software
- Proprietary cross-platform software
- Cross-platform software
- Windows software
- Linux science software
- Mac OS X software
- PlayStation 3 software
- 2000 introductions
