Genetic code: Difference between revisions
Jezhotwells (talk | contribs) |
m →Degeneracy of the genetic code: typo |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
== Cracking the genetic code == |
== Cracking the genetic code == |
||
[[Image:GeneticCode21-version-2.svg|thumb|The genetic code]] |
[[Image:GeneticCode21-version-2.svg|thumb|The genetic code]] |
||
After the structure of DNA was deciphered by [[James D. Watson|James Watson]], [[Francis Crick]], [[Maurice Wilkins]] and [[Rosalind Franklin]], serious efforts to understand the nature of the encoding of proteins began. [[George Gamow]] postulated that a three-letter code must be employed to encode the 20 standard [[amino acid]]s used by living cells to encode proteins (because 3 is the smallest integer n such that 4<sup>n</sup> is at least 20).{{ |
After the structure of DNA was deciphered by [[James D. Watson|James Watson]], [[Francis Crick]], [[Maurice Wilkins]] and [[Rosalind Franklin]], serious efforts to understand the nature of the encoding of proteins began. [[George Gamow]] postulated that a three-letter code must be employed to encode the 20 standard [[amino acid]]s used by living cells to encode proteins (because 3 is the smallest integer n such that 4<sup>n</sup> is at least 20).<ref name="isbn0-465-09138-5">{{cite book | author = Crick, Francis | authorlink = | editor = | others = | title = What mad pursuit: a personal view of scientific discovery | edition = | language = | publisher = Basic Books | location = New York | year = 1988 | origyear = | pages = 89-101 | quote = | isbn = 0-465-09138-5 | oclc = | doi = | url = | accessdate = | chapter = Chapter 8: The genetic code }}</ref> |
||
The fact that codons did consist of three DNA bases was first demonstrated in the [[Crick, Brenner et al. experiment]]. The first elucidation of a codon was done by [[Marshall Nirenberg]] and [[Heinrich J. Matthaei]] in 1961 at the [[National Institutes of Health]]. They used a [[cell-free system]] to [[translation (biology)|translate]] a poly-uracil RNA sequence (or UUUUU... in biochemical terms) and discovered that the [[polypeptide]] that they had synthesized consisted of only the amino acid [[phenylalanine]]. They thereby deduced from this poly-phenylalanine that the codon UUU specified the amino-acid phenylalanine. Extending this work, Nirenberg and [[Philip Leder]] were able to elucidate the triplet nature of the genetic code and allowed the codons of the standard genetic code to be deciphered. In these experiments, various combinations of mRNA were passed through a filter which contained ribosomes. Unique triplets promoted the binding of specific tRNAs to the ribosome. Leder and Nirenberg were able to determine the sequences of 54 out of 64 codons.{{ |
The fact that codons did consist of three DNA bases was first demonstrated in the [[Crick, Brenner et al. experiment]]. The first elucidation of a codon was done by [[Marshall Nirenberg]] and [[Heinrich J. Matthaei]] in 1961 at the [[National Institutes of Health]]. They used a [[cell-free system]] to [[translation (biology)|translate]] a poly-uracil RNA sequence (or UUUUU... in biochemical terms) and discovered that the [[polypeptide]] that they had synthesized consisted of only the amino acid [[phenylalanine]]. They thereby deduced from this poly-phenylalanine that the codon UUU specified the amino-acid phenylalanine. Extending this work, Nirenberg and [[Philip Leder]] were able to elucidate the triplet nature of the genetic code and allowed the codons of the standard genetic code to be deciphered. In these experiments, various combinations of mRNA were passed through a filter which contained ribosomes. Unique triplets promoted the binding of specific tRNAs to the ribosome. Leder and Nirenberg were able to determine the sequences of 54 out of 64 codons.<ref name="pmid5330357">{{cite journal | author = Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, Rottman F, O'Neal C | title = RNA codewords and protein synthesis, VII. On the general nature of the RNA code | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 53 | issue = 5 | pages = 1161–8 | year = 1965 | month = May | pmid = 5330357 | pmc = 301388 | doi = | url = | issn = }}</ref> |
||
Subsequent work by [[Har Gobind Khorana]] identified the rest of the code, and shortly thereafter [[Robert W. Holley]] determined the structure of [[transfer RNA]], the adapter molecule that facilitates translation. This work was based upon earlier studies by [[Severo Ochoa]], who received the Nobel prize in 1959 for his work on the enzymology of RNA synthesis. In 1968, Khorana, Holley and Nirenberg received the [[Nobel Prize]] in Physiology or Medicine for their work.{{ |
Subsequent work by [[Har Gobind Khorana]] identified the rest of the code, and shortly thereafter [[Robert W. Holley]] determined the structure of [[transfer RNA]], the adapter molecule that facilitates translation. This work was based upon earlier studies by [[Severo Ochoa]], who received the Nobel prize in 1959 for his work on the enzymology of RNA synthesis.<ref name = "Nobel_1959">{{cite press release | url = http://nobelprize.org/nobel_prizes/medicine/laureates/1959/index.html | title = The Nobel Prize in Physiology or Medicine 1959 | publisher = The Royal Swedish Academy of Science | date = 1959 | accessdate = 2010-02-27 }}</ref> In 1968, Khorana, Holley and Nirenberg received the [[Nobel Prize]] in Physiology or Medicine for their work.<ref name = "Nobel_1968">{{cite press release | url = http://nobelprize.org/nobel_prizes/medicine/laureates/1968/index.html | title = The Nobel Prize in Physiology or Medicine 1968 | publisher = The Royal Swedish Academy of Science | date = 1968 | accessdate = 2010-02-27 }}</ref> |
||
== Transfer of information via the genetic code == |
== Transfer of information via the genetic code == |
||
The [[genome]] of an [[organism]] is inscribed in [[DNA]], or in the case of some viruses, [[RNA]]. The portion of the genome that codes for a protein or an RNA is referred to as a [[gene]]. Those genes that code for proteins are composed of tri-nucleotide units called '''codons''', each coding for a single amino acid. Each nucleotide sub-unit consists of a [[phosphate]], [[deoxyribose]] sugar and one of the 4 nitrogenous [[nucleotide]] bases. The [[purine]] bases [[adenine]] ('''A''') and [[guanine]] ('''G''') are larger and consist of two aromatic rings. The [[pyrimidine]] bases [[cytosine]] ('''C''') and [[thymine]] ('''T''') are smaller and consist of only one aromatic ring. In the double-helix configuration, two strands of DNA are joined to each other by hydrogen bonds in an arrangement known as [[base pair]]ing. These bonds almost always form between an adenine base on one strand and a thymine on the other strand and between a cytosine base on one strand and a guanine base on the other. This means that the number of A and T residues will be the same in a given double helix, as will the number of G and C residues. In RNA, thymine ('''T''') is replaced by [[uracil]] ('''U'''), and the deoxyribose is substituted by [[ribose]].{{ |
The [[genome]] of an [[organism]] is inscribed in [[DNA]], or in the case of some viruses, [[RNA]]. The portion of the genome that codes for a protein or an RNA is referred to as a [[gene]]. Those genes that code for proteins are composed of tri-nucleotide units called '''codons''', each coding for a single amino acid. Each nucleotide sub-unit consists of a [[phosphate]], [[deoxyribose]] sugar and one of the 4 nitrogenous [[nucleotide]] bases. The [[purine]] bases [[adenine]] ('''A''') and [[guanine]] ('''G''') are larger and consist of two aromatic rings. The [[pyrimidine]] bases [[cytosine]] ('''C''') and [[thymine]] ('''T''') are smaller and consist of only one aromatic ring. In the double-helix configuration, two strands of DNA are joined to each other by hydrogen bonds in an arrangement known as [[base pair]]ing. These bonds almost always form between an adenine base on one strand and a thymine on the other strand and between a cytosine base on one strand and a guanine base on the other. This means that the number of A and T residues will be the same in a given double helix, as will the number of G and C residues. In RNA, thymine ('''T''') is replaced by [[uracil]] ('''U'''), and the deoxyribose is substituted by [[ribose]].<ref name="MBG">{{cite book | author = Losick, Richard; Watson, James D.; Tania A. Baker; Bell, Stephen; Gann, Alexander; Levine, Michael W. | authorlink = | editor = | others = | title = Molecular Biology of the Gene | edition = | language = | publisher = Pearson/Benjamin Cummings | location = San Francisco | year = 2008 | origyear = | pages = | quote = | isbn = 0-8053-9592-X | oclc = | doi = | url = | accessdate = }}</ref>{{rp|Chp 6}} |
||
Each protein-coding gene is [[transcription (genetics)|transcribed]] into a template molecule of the related polymer RNA, known as [[messenger RNA]] or mRNA. This, in turn, is [[translation (genetics)|translated]] on the [[ribosome]] into an [[amino acid]] chain or [[peptide|polypeptide]]. The process of translation requires [[transfer RNA]]s specific for individual amino acids with the amino acids [[covalent]]ly attached to them, [[guanosine triphosphate]] as an energy source, and a number of translation factors. tRNAs have [[anticodons]] complementary to the codons in mRNA and can be "charged" covalently with amino acids at their 3' terminal CCA ends. Individual tRNAs are charged with specific amino acids by enzymes known as [[aminoacyl tRNA synthetase]]s, which have high specificity for both their cognate amino acids and tRNAs. The high specificity of these enzymes is a major reason why the fidelity of protein translation is maintained.{{ |
Each protein-coding gene is [[transcription (genetics)|transcribed]] into a template molecule of the related polymer RNA, known as [[messenger RNA]] or mRNA. This, in turn, is [[translation (genetics)|translated]] on the [[ribosome]] into an [[amino acid]] chain or [[peptide|polypeptide]].<ref name="MBG"/>{{rp|Chp 12}} The process of translation requires [[transfer RNA]]s specific for individual amino acids with the amino acids [[covalent]]ly attached to them, [[guanosine triphosphate]] as an energy source, and a number of translation factors. tRNAs have [[anticodons]] complementary to the codons in mRNA and can be "charged" covalently with amino acids at their 3' terminal CCA ends. Individual tRNAs are charged with specific amino acids by enzymes known as [[aminoacyl tRNA synthetase]]s, which have high specificity for both their cognate amino acids and tRNAs. The high specificity of these enzymes is a major reason why the fidelity of protein translation is maintained.<ref name="MBG"/>{{rp|Chp 14}} |
||
There are 4³ = 64 different codon combinations possible with a triplet codon of three nucleotides; all 64 codons are assigned for either amino acids or stop signals during translation. If, for example, an RNA sequence, UUUAAACCC is considered and the '''[[reading frames|reading-frame]]''' starts with the first U (by convention, [[DNA#Strand direction|5' to 3']]), there are three codons, namely, UUU, AAA and CCC, each of which specifies one amino acid. This RNA sequence will be translated into an amino acid sequence, three amino acids long. A comparison may be made with [[computer science]], where the codon is similar to a [[word (computing)|word]], which is the standard "chunk" for handling data (like one amino acid of a protein), and a nucleotide is similar to a [[bit]], in that it is the smallest unit.{{ |
There are 4³ = 64 different codon combinations possible with a triplet codon of three nucleotides; all 64 codons are assigned for either amino acids or stop signals during translation. If, for example, an RNA sequence, UUUAAACCC is considered and the '''[[reading frames|reading-frame]]''' starts with the first U (by convention, [[DNA#Strand direction|5' to 3']]), there are three codons, namely, UUU, AAA and CCC, each of which specifies one amino acid. This RNA sequence will be translated into an amino acid sequence, three amino acids long. A comparison may be made with [[computer science]], where the codon is similar to a [[word (computing)|word]], which is the standard "chunk" for handling data (like one amino acid of a protein), and a nucleotide is similar to a [[bit]], in that it is the smallest unit.<ref name="MBG"/>{{rp|Chp 15}} |
||
The standard genetic code is shown in the following tables. Table 1 shows what amino acid each of the 64 codons specifies. Table 2 shows what codons specify each of the 20 standard amino acids involved in translation. These are called forward and reverse codon tables, respectively. For example, the codon AAU represents the amino acid [[asparagine]], and UGU and UGC represent [[cysteine]] (standard three-letter designations, Asn and Cys, respectively).{{ |
The standard genetic code is shown in the following tables. Table 1 shows what amino acid each of the 64 codons specifies. Table 2 shows what codons specify each of the 20 standard amino acids involved in translation. These are called forward and reverse codon tables, respectively. For example, the codon AAU represents the amino acid [[asparagine]], and UGU and UGC represent [[cysteine]] (standard three-letter designations, Asn and Cys, respectively).<ref name="MBG"/>{{rp|Chp 15}} |
||
== RNA codon table ==<!-- This section is linked from [[Amino acid]] --> |
== RNA codon table ==<!-- This section is linked from [[Amino acid]] --> |
||
| Line 193: | Line 193: | ||
== Salient features == |
== Salient features == |
||
===Sequence reading frame=== |
===Sequence reading frame=== |
||
A [[codon]] is defined by the initial nucleotide from which translation starts. For example, the string GGGAAACCC, if read from the first position, contains the codons GGG, AAA and CCC; and, if read from the second position, it contains the codons GGA and AAC; if read starting from the third position, GAA and ACC. Partial codons have been ignored in this example. Every sequence can thus be read in three '''[[reading frames]]''', each of which will produce a different amino acid sequence (in the given example, Gly-Lys-Pro, Gly-Asn, or Glu-Thr, respectively). With double-stranded DNA there are six possible [[reading frames]], three in the forward orientation on one strand and three reverse (on the opposite strand).{{ |
A [[codon]] is defined by the initial nucleotide from which translation starts. For example, the string GGGAAACCC, if read from the first position, contains the codons GGG, AAA and CCC; and, if read from the second position, it contains the codons GGA and AAC; if read starting from the third position, GAA and ACC. Partial codons have been ignored in this example. Every sequence can thus be read in three '''[[reading frames]]''', each of which will produce a different amino acid sequence (in the given example, Gly-Lys-Pro, Gly-Asn, or Glu-Thr, respectively). With double-stranded DNA there are six possible [[reading frames]], three in the forward orientation on one strand and three reverse (on the opposite strand).<ref name="genetics_ dictionary"/>{{rp|330}} |
||
The actual frame in which a protein sequence is translated is defined by a '''[[start codon]]''', usually the first AUG codon in the mRNA sequence. Mutations that disrupt the reading frame by insertions or deletions of a non-multiple of 3 nucleotide bases are known as [[frameshift mutation]]s. These mutations may impair the function of the resulting protein, if it is formed, and are thus rare in ''[[in vivo]]'' protein-coding sequences. Often such misformed proteins are targeted for [[proteolytic]] degradation. In addition, a frame shift mutation is very likely to cause a [[stop codon]] to be read, which truncates the creation of the protein.<ref>{{cite journal |author=Isbrandt D, Hopwood JJ, von Figura K, Peters C |title=Two novel frameshift mutations causing premature stop codons in a patient with the severe form of Maroteaux-Lamy syndrome |journal=Hum. Mutat. |volume=7 |issue=4 |pages=361–3 |year=1996 |pmid=8723688 |doi=10.1002/(SICI)1098-1004(1996)7:4<361::AID-HUMU12>3.0.CO;2-0 |doi_brokendate=2009-07-09 }}</ref> One reason for the rareness of frame-shifted mutations' being inherited is that, if the protein being translated is essential for growth under the selective pressures the organism faces, absence of a functional protein may cause lethality before the organism is viable.{{ |
The actual frame in which a protein sequence is translated is defined by a '''[[start codon]]''', usually the first AUG codon in the mRNA sequence. Mutations that disrupt the reading frame by insertions or deletions of a non-multiple of 3 nucleotide bases are known as [[frameshift mutation]]s. These mutations may impair the function of the resulting protein, if it is formed, and are thus rare in ''[[in vivo]]'' protein-coding sequences. Often such misformed proteins are targeted for [[proteolytic]] degradation. In addition, a frame shift mutation is very likely to cause a [[stop codon]] to be read, which truncates the creation of the protein.<ref>{{cite journal |author=Isbrandt D, Hopwood JJ, von Figura K, Peters C |title=Two novel frameshift mutations causing premature stop codons in a patient with the severe form of Maroteaux-Lamy syndrome |journal=Hum. Mutat. |volume=7 |issue=4 |pages=361–3 |year=1996 |pmid=8723688 |doi=10.1002/(SICI)1098-1004(1996)7:4<361::AID-HUMU12>3.0.CO;2-0 |doi_brokendate=2009-07-09 }}</ref> One reason for the rareness of frame-shifted mutations' being inherited is that, if the protein being translated is essential for growth under the selective pressures the organism faces, absence of a functional protein may cause lethality before the organism is viable.<ref name="pmid8444142">{{cite journal | author = Crow JF | title = How much do we know about spontaneous human mutation rates? | journal = Environ. Mol. Mutagen. | volume = 21 | issue = 2 | pages = 122–9 | year = 1993 | pmid = 8444142 | doi = | url = | issn = }}</ref> |
||
=== Start/stop codons === |
=== Start/stop codons === |
||
| Line 206: | Line 207: | ||
[[File:Notable mutations.svg|300px|thumb|Selection of notable [[mutation]]s.<ref>References for the image are found in Wikimedia Commons page at: [[Commons:File:Notable mutations.svg#References]].</ref>]]<!-- EXPANSION OF THE IMAGE WITH MORE EXAMPLES IS EXPECTED (see its discussion page)--> |
[[File:Notable mutations.svg|300px|thumb|Selection of notable [[mutation]]s.<ref>References for the image are found in Wikimedia Commons page at: [[Commons:File:Notable mutations.svg#References]].</ref>]]<!-- EXPANSION OF THE IMAGE WITH MORE EXAMPLES IS EXPECTED (see its discussion page)--> |
||
[[Frameshift mutations]] altering the sequence reading frame, and [[nonsense mutation]]s causing a stop codon are examples of [[point mutation]]s. In addition, there may be [[missense mutation]]s that cause exchange of one amino acid for another. Clinically important [[missense mutations]] generally change the properties of the coded amino acid residue between being basic, acidic polar or nonpolar, while [[nonsense mutations]] result in a [[stop codon]].{{ |
[[Frameshift mutations]] altering the sequence reading frame, and [[nonsense mutation]]s causing a stop codon are examples of [[point mutation]]s. In addition, there may be [[missense mutation]]s that cause exchange of one amino acid for another. Clinically important [[missense mutations]] generally change the properties of the coded amino acid residue between being basic, acidic polar or nonpolar, while [[nonsense mutations]] result in a [[stop codon]].<ref name="genetics_ dictionary">{{cite book | author = Pamela K. Mulligan; King, Robert C.; Stansfield, William D. | authorlink = | editor = | others = | title = A dictionary of genetics | edition = | language = | publisher = Oxford University Press | location = Oxford [Oxfordshire] | year = 2006 | origyear = | pages = 608 | quote = | isbn = 0-19-530761-5 | oclc = | doi = | url = | accessdate = }}</ref>{{rp|266}} |
||
=== Degeneracy of the genetic code ===<!-- This section is linked from [[Neutral theory of molecular evolution]] --> |
=== Degeneracy of the genetic code ===<!-- This section is linked from [[Neutral theory of molecular evolution]] --> |
||
The genetic code has redundancy but no ambiguity (see the [[#RNA codon table|codon tables]] above for the full correlation). For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither of them specifies any other amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example the amino acid [[glutamic acid]] is specified by GAA and GAG codons (difference in the third position), the amino acid [[leucine]] is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position), while the amino acid [[serine]] is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second or third position).{{ |
The genetic code has redundancy but no ambiguity (see the [[#RNA codon table|codon tables]] above for the full correlation). For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither of them specifies any other amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example the amino acid [[glutamic acid]] is specified by GAA and GAG codons (difference in the third position), the amino acid [[leucine]] is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position), while the amino acid [[serine]] is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second or third position).<ref name="MBG"/>{{rp|Chp 15}} |
||
A position of a codon is said to be a '''fourfold degenerate site''' if any nucleotide at this position specifies the same amino acid. For example, the third position of the [[glycine]] codons (GGA, GGG, GGC, GGU) is a fourfold degenerate site, because all nucleotide substitutions at this site are synonymous; i.e., they do not change the amino acid. Only the third positions of some codons may be fourfold degenerate.{{ |
A position of a codon is said to be a '''fourfold degenerate site''' if any nucleotide at this position specifies the same amino acid. For example, the third position of the [[glycine]] codons (GGA, GGG, GGC, GGU) is a fourfold degenerate site, because all nucleotide substitutions at this site are synonymous; i.e., they do not change the amino acid. Only the third positions of some codons may be fourfold degenerate.<ref name="MBG"/>{{rp|Chp 15}} |
||
A position of a codon is said to be a '''twofold degenerate site''' if only two of four possible nucleotides at this position specify the same amino acid. For example, the third position of the [[glutamic acid]] codons (GAA, GAG) is a twofold degenerate site. In twofold degenerate sites, the equivalent nucleotides are always either two [[purine]]s (A/G) or two [[pyrimidine]]s (C/U), so only transversional substitutions (purine to pyrimidine or pyrimidine to purine) in twofold degenerate sites are nonsynonymous.{{ |
A position of a codon is said to be a '''twofold degenerate site''' if only two of four possible nucleotides at this position specify the same amino acid. For example, the third position of the [[glutamic acid]] codons (GAA, GAG) is a twofold degenerate site. In twofold degenerate sites, the equivalent nucleotides are always either two [[purine]]s (A/G) or two [[pyrimidine]]s (C/U), so only transversional substitutions (purine to pyrimidine or pyrimidine to purine) in twofold degenerate sites are nonsynonymous.<ref name="MBG"/>{{rp|Chp 15}} |
||
A position of a codon is said to be a '''non-degenerate site''' if any mutation at this position results in amino acid substitution. There is only one '''threefold degenerate site''' where changing three of the four nucleotides may have no effect on the amino acid (depending on what it is changed to), while changing the fourth possible nucleotide always results in an amino acid substitution. This is the third position of an [[isoleucine]] codon: AUU, AUC, or AUA all encode isoleucine, but AUG encodes [[methionine]]. In computation this position is often treated as a twofold degenerate site.{{ |
A position of a codon is said to be a '''non-degenerate site''' if any mutation at this position results in amino acid substitution. There is only one '''threefold degenerate site''' where changing three of the four nucleotides may have no effect on the amino acid (depending on what it is changed to), while changing the fourth possible nucleotide always results in an amino acid substitution. This is the third position of an [[isoleucine]] codon: AUU, AUC, or AUA all encode isoleucine, but AUG encodes [[methionine]]. In computation this position is often treated as a twofold degenerate site.<ref name="MBG"/>{{rp|Chp 15}} |
||
There are three amino acids encoded by six different codons: [[serine]], [[leucine]], [[arginine]]. Only two amino acids are specified by a single codon; one of these is the amino-acid [[methionine]], specified by the codon AUG, which also specifies the start of translation; the other is [[tryptophan]], specified by the codon UGG. |
There are three amino acids encoded by six different codons: [[serine]], [[leucine]], [[arginine]]. Only two amino acids are specified by a single codon; one of these is the amino-acid [[methionine]], specified by the codon AUG, which also specifies the start of translation; the other is [[tryptophan]], specified by the codon UGG. |
||
The degeneracy of the genetic code is what accounts for the existence of [[synonymous mutation]]s.{{ |
The degeneracy of the genetic code is what accounts for the existence of [[synonymous mutation]]s.<ref name="MBG"/>{{rp|Chp 15}} |
||
Degeneracy results because a triplet code designates 20 amino acids and a stop codon. Because there are four bases, triplet codons are required to produce at least 21 different codes. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required, then 4³ gives 64 possible codons, meaning that some degeneracy must exist.{{ |
Degeneracy results because a triplet code designates 20 amino acids and a stop codon. Because there are four bases, triplet codons are required to produce at least 21 different codes. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required, then 4³ gives 64 possible codons, meaning that some degeneracy must exist.<ref name="MBG"/>{{rp|Chp 15}} |
||
These properties of the genetic code make it more fault-tolerant for [[point mutation]]s. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although [[codon usage bias]] restricts this in practice in many organisms; twofold degenerate codons can tolerate one out of the three possible point mutations at the third position. Since [[transition (genetics)|transition]] mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than [[transversion]] (purine to pyrimidine or vice-versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.{{ |
These properties of the genetic code make it more fault-tolerant for [[point mutation]]s. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although [[codon usage bias]] restricts this in practice in many organisms; twofold degenerate codons can tolerate one out of the three possible point mutations at the third position. Since [[transition (genetics)|transition]] mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than [[transversion]] (purine to pyrimidine or vice-versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.<ref name="MBG"/>{{rp|Chp 15}} |
||
[[Image:Genetic Code Bias 2.svg|thumb|Grouping of [[codon]]s by amino acid residue molar volume and [[hydropathy]].]] |
[[Image:Genetic Code Bias 2.svg|thumb|Grouping of [[codon]]s by amino acid residue molar volume and [[hydropathy]].]] |
||
| Line 227: | Line 228: | ||
A practical consequence of redundancy is that some errors in the genetic code only cause a silent mutation or an error that would not affect the protein because the [[hydrophilic]]ity or [[hydrophobic]]ity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathy; NAN encodes average size hydrophilic residues.<ref>{{cite book |author=Yang ''et al.'' |editor=Michel-Beyerle, M. E. |title=Reaction centers of photosynthetic bacteria: Feldafing-II-Meeting |publisher=Springer-Verlag |location=Berlin |year=1990 |pages=209–18 |isbn=3-540-53420-2 |volume=6}}</ref><ref>{{cite journal |author=Füllen G, Youvan DC |title=Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering |journal=Complexity International |volume=1 |year=1994 |url=http://www.complexity.org.au/ci/vol01/fullen01/html/}}</ref> These tendencies may result from that the [[aminoacyl tRNA synthetases]] related the such codons share a common ancestry. |
A practical consequence of redundancy is that some errors in the genetic code only cause a silent mutation or an error that would not affect the protein because the [[hydrophilic]]ity or [[hydrophobic]]ity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathy; NAN encodes average size hydrophilic residues.<ref>{{cite book |author=Yang ''et al.'' |editor=Michel-Beyerle, M. E. |title=Reaction centers of photosynthetic bacteria: Feldafing-II-Meeting |publisher=Springer-Verlag |location=Berlin |year=1990 |pages=209–18 |isbn=3-540-53420-2 |volume=6}}</ref><ref>{{cite journal |author=Füllen G, Youvan DC |title=Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering |journal=Complexity International |volume=1 |year=1994 |url=http://www.complexity.org.au/ci/vol01/fullen01/html/}}</ref> These tendencies may result from that the [[aminoacyl tRNA synthetases]] related the such codons share a common ancestry. |
||
Even so, single point mutations can still cause dysfunctional proteins. For example, a mutated [[hemoglobin]] gene causes [[sickle-cell disease]]. In the mutant hemoglobin a hydrophilic [[glutamate]] (Glu) is substituted by the hydrophobic [[valine]] (Val), that is, GAA or GAG becomes GUA or GUG. The substitution of glutamate by valine reduces the solubility of [[Beta globulins|β-globin]] which causes [[hemoglobin]] to form linear polymers linked by the hydrophobic interaction between the valine groups causing sickle-cell deformation of erythrocytes. Sickle-cell disease is generally not caused by a ''de novo'' [[mutation]]. Rather it is selected for in [[malaria]]l regions (in a way similar to [[thalassemia]]), as [[Zygosity|heterozygous]] people have some resistance to the malarial ''[[Plasmodium]]'' parasite ([[heterozygote advantage]]).{{ |
Even so, single point mutations can still cause dysfunctional proteins. For example, a mutated [[hemoglobin]] gene causes [[sickle-cell disease]]. In the mutant hemoglobin a hydrophilic [[glutamate]] (Glu) is substituted by the hydrophobic [[valine]] (Val), that is, GAA or GAG becomes GUA or GUG. The substitution of glutamate by valine reduces the solubility of [[Beta globulins|β-globin]] which causes [[hemoglobin]] to form linear polymers linked by the hydrophobic interaction between the valine groups causing sickle-cell deformation of erythrocytes. Sickle-cell disease is generally not caused by a ''de novo'' [[mutation]]. Rather it is selected for in [[malaria]]l regions (in a way similar to [[thalassemia]]), as [[Zygosity|heterozygous]] people have some resistance to the malarial ''[[Plasmodium]]'' parasite ([[heterozygote advantage]]).<ref name="pmid14962356">{{cite journal | author = Hebbel RP | title = Sickle hemoglobin instability: a mechanism for malarial protection | journal = Redox Rep. | volume = 8 | issue = 5 | pages = 238–40 | year = 2003 | pmid = 14962356 | doi = 10.1179/135100003225002826 | url = | issn = }}</ref> |
||
These variable codes for amino acids are allowed because of modified bases in the first base of the [[anticodon]] of the tRNA, and the base-pair formed is called a [[wobble base pair]]. The modified bases include [[inosine]] and the Non-Watson-Crick U-G basepair.{{ |
These variable codes for amino acids are allowed because of modified bases in the first base of the [[anticodon]] of the tRNA, and the base-pair formed is called a [[wobble base pair]]. The modified bases include [[inosine]] and the Non-Watson-Crick U-G basepair.<ref name="pmid11256617">{{cite journal | author = Varani G, McClain WH | title = The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems | journal = EMBO Rep. | volume = 1 | issue = 1 | pages = 18–23 | year = 2000 | month = July | pmid = 11256617 | pmc = 1083677 | doi = 10.1093/embo-reports/kvd001 | url = | issn = }}</ref> |
||
== Variations to the standard genetic code == |
== Variations to the standard genetic code == |
||
Revision as of 09:07, 27 February 2010

The genetic code is the set of rules by which information encoded in genetic material (DNA or mRNA sequences) is translated into proteins (amino acid sequences) by living cells. The code defines a mapping between tri-nucleotide sequences, called codons, and amino acids. A triplet codon in a nucleic acid sequence usually specifies a single amino acid (though in some cases the same codon triplet in different locations can code unambiguously for two different amino acids, the correct choice at each location being determined by context).[1] Because the vast majority of genes are encoded with exactly the same code (see the RNA codon table), this particular code is often referred to as the canonical or standard genetic code, or simply the genetic code, though in fact there are many variant codes. Thus the canonical genetic code is not universal. For example, in humans, protein synthesis in mitochondria relies on a genetic code that varies from the canonical code.
It is important to know that not all genetic information is stored using the genetic code. All organisms' DNA contain regulatory sequences, intergenic segments, and chromosomal structural areas that can contribute greatly to phenotype but operate using distinct sets of rules that may or may not be as straightforward as the codon-to-amino acid paradigm that usually underlies the genetic code (see epigenetics).
Cracking the genetic code

After the structure of DNA was deciphered by James Watson, Francis Crick, Maurice Wilkins and Rosalind Franklin, serious efforts to understand the nature of the encoding of proteins began. George Gamow postulated that a three-letter code must be employed to encode the 20 standard amino acids used by living cells to encode proteins (because 3 is the smallest integer n such that 4n is at least 20).[2]
The fact that codons did consist of three DNA bases was first demonstrated in the Crick, Brenner et al. experiment. The first elucidation of a codon was done by Marshall Nirenberg and Heinrich J. Matthaei in 1961 at the National Institutes of Health. They used a cell-free system to translate a poly-uracil RNA sequence (or UUUUU... in biochemical terms) and discovered that the polypeptide that they had synthesized consisted of only the amino acid phenylalanine. They thereby deduced from this poly-phenylalanine that the codon UUU specified the amino-acid phenylalanine. Extending this work, Nirenberg and Philip Leder were able to elucidate the triplet nature of the genetic code and allowed the codons of the standard genetic code to be deciphered. In these experiments, various combinations of mRNA were passed through a filter which contained ribosomes. Unique triplets promoted the binding of specific tRNAs to the ribosome. Leder and Nirenberg were able to determine the sequences of 54 out of 64 codons.[3]
Subsequent work by Har Gobind Khorana identified the rest of the code, and shortly thereafter Robert W. Holley determined the structure of transfer RNA, the adapter molecule that facilitates translation. This work was based upon earlier studies by Severo Ochoa, who received the Nobel prize in 1959 for his work on the enzymology of RNA synthesis.[4] In 1968, Khorana, Holley and Nirenberg received the Nobel Prize in Physiology or Medicine for their work.[5]
Transfer of information via the genetic code
The genome of an organism is inscribed in DNA, or in the case of some viruses, RNA. The portion of the genome that codes for a protein or an RNA is referred to as a gene. Those genes that code for proteins are composed of tri-nucleotide units called codons, each coding for a single amino acid. Each nucleotide sub-unit consists of a phosphate, deoxyribose sugar and one of the 4 nitrogenous nucleotide bases. The purine bases adenine (A) and guanine (G) are larger and consist of two aromatic rings. The pyrimidine bases cytosine (C) and thymine (T) are smaller and consist of only one aromatic ring. In the double-helix configuration, two strands of DNA are joined to each other by hydrogen bonds in an arrangement known as base pairing. These bonds almost always form between an adenine base on one strand and a thymine on the other strand and between a cytosine base on one strand and a guanine base on the other. This means that the number of A and T residues will be the same in a given double helix, as will the number of G and C residues. In RNA, thymine (T) is replaced by uracil (U), and the deoxyribose is substituted by ribose.[6]: Chp 6
Each protein-coding gene is transcribed into a template molecule of the related polymer RNA, known as messenger RNA or mRNA. This, in turn, is translated on the ribosome into an amino acid chain or polypeptide.[6]: Chp 12 The process of translation requires transfer RNAs specific for individual amino acids with the amino acids covalently attached to them, guanosine triphosphate as an energy source, and a number of translation factors. tRNAs have anticodons complementary to the codons in mRNA and can be "charged" covalently with amino acids at their 3' terminal CCA ends. Individual tRNAs are charged with specific amino acids by enzymes known as aminoacyl tRNA synthetases, which have high specificity for both their cognate amino acids and tRNAs. The high specificity of these enzymes is a major reason why the fidelity of protein translation is maintained.[6]: Chp 14
There are 4³ = 64 different codon combinations possible with a triplet codon of three nucleotides; all 64 codons are assigned for either amino acids or stop signals during translation. If, for example, an RNA sequence, UUUAAACCC is considered and the reading-frame starts with the first U (by convention, 5' to 3'), there are three codons, namely, UUU, AAA and CCC, each of which specifies one amino acid. This RNA sequence will be translated into an amino acid sequence, three amino acids long. A comparison may be made with computer science, where the codon is similar to a word, which is the standard "chunk" for handling data (like one amino acid of a protein), and a nucleotide is similar to a bit, in that it is the smallest unit.[6]: Chp 15
The standard genetic code is shown in the following tables. Table 1 shows what amino acid each of the 64 codons specifies. Table 2 shows what codons specify each of the 20 standard amino acids involved in translation. These are called forward and reverse codon tables, respectively. For example, the codon AAU represents the amino acid asparagine, and UGU and UGC represent cysteine (standard three-letter designations, Asn and Cys, respectively).[6]: Chp 15
RNA codon table
| nonpolar | polar | basic | acidic | (stop codon) |
| 2nd base | |||||
|---|---|---|---|---|---|
| U | C | A | G | ||
| 1st base |
U | UUU (Phe/F) Phenylalanine UUC (Phe/F) Phenylalanine |
UCU (Ser/S) Serine UCC (Ser/S) Serine |
UAU (Tyr/Y) Tyrosine UAC (Tyr/Y) Tyrosine |
UGU (Cys/C) Cysteine UGC (Cys/C) Cysteine |
| UUA (Leu/L) Leucine | UCA (Ser/S) Serine | UAA Ochre (Stop) | UGA Opal (Stop) | ||
| UUG (Leu/L) Leucine | UCG (Ser/S) Serine | UAG Amber (Stop) | UGG (Trp/W) Tryptophan | ||
| C | CUU (Leu/L) Leucine CUC (Leu/L) Leucine |
CCU (Pro/P) Proline CCC (Pro/P) Proline |
CAU (His/H) Histidine CAC (His/H) Histidine |
CGU (Arg/R) Arginine CGC (Arg/R) Arginine | |
| CUA (Leu/L) Leucine CUG (Leu/L) Leucine |
CCA (Pro/P) Proline CCG (Pro/P) Proline |
CAA (Gln/Q) Glutamine
CAG (Gln/Q) Glutamine |
CGA (Arg/R) Arginine CGG (Arg/R) Arginine | ||
| A | AUU (Ile/I) Isoleucine AUC (Ile/I) Isoleucine |
ACU (Thr/T) Threonine ACC (Thr/T) Threonine |
AAU (Asn/N) Asparagine AAC (Asn/N) Asparagine |
AGU (Ser/S) Serine AGC (Ser/S) Serine | |
| AUA (Ile/I) Isoleucine | ACA (Thr/T) Threonine | AAA (Lys/K) Lysine | AGA (Arg/R) Arginine | ||
| AUG[A] (Met/M) Methionine |
ACG (Thr/T) Threonine | AAG (Lys/K) Lysine | AGG (Arg/R) Arginine | ||
| G | GUU (Val/V) Valine GUC (Val/V) Valine |
GCU (Ala/A) Alanine GCC (Ala/A) Alanine |
GAU (Asp/D) Aspartic acid GAC (Asp/D) Aspartic acid |
GGU (Gly/G) Glycine GGC (Gly/G) Glycine | |
| GUA (Val/V) Valine GUG (Val/V) Valine |
GCA (Ala/A) Alanine GCG (Ala/A) Alanine |
GAA (Glu/E) Glutamic acid GAG (Glu/E) Glutamic acid |
GGA (Gly/G) Glycine GGG (Gly/G) Glycine | ||
- A The codon AUG both codes for methionine and serves as an initiation site: the first AUG in an mRNA's coding region is where translation into protein begins.[7]
| Ala/A | GCU, GCC, GCA, GCG | Leu/L | UUA, UUG, CUU, CUC, CUA, CUG |
|---|---|---|---|
| Arg/R | CGU, CGC, CGA, CGG, AGA, AGG | Lys/K | AAA, AAG |
| Asn/N | AAU, AAC | Met/M | AUG |
| Asp/D | GAU, GAC | Phe/F | UUU, UUC |
| Cys/C | UGU, UGC | Pro/P | CCU, CCC, CCA, CCG |
| Gln/Q | CAA, CAG | Ser/S | UCU, UCC, UCA, UCG, AGU, AGC |
| Glu/E | GAA, GAG | Thr/T | ACU, ACC, ACA, ACG |
| Gly/G | GGU, GGC, GGA, GGG | Trp/W | UGG |
| His/H | CAU, CAC | Tyr/Y | UAU, UAC |
| Ile/I | AUU, AUC, AUA | Val/V | GUU, GUC, GUA, GUG |
| START | AUG | STOP | UAA, UGA, UAG |
Salient features
Sequence reading frame
A codon is defined by the initial nucleotide from which translation starts. For example, the string GGGAAACCC, if read from the first position, contains the codons GGG, AAA and CCC; and, if read from the second position, it contains the codons GGA and AAC; if read starting from the third position, GAA and ACC. Partial codons have been ignored in this example. Every sequence can thus be read in three reading frames, each of which will produce a different amino acid sequence (in the given example, Gly-Lys-Pro, Gly-Asn, or Glu-Thr, respectively). With double-stranded DNA there are six possible reading frames, three in the forward orientation on one strand and three reverse (on the opposite strand).[8]: 330
The actual frame in which a protein sequence is translated is defined by a start codon, usually the first AUG codon in the mRNA sequence. Mutations that disrupt the reading frame by insertions or deletions of a non-multiple of 3 nucleotide bases are known as frameshift mutations. These mutations may impair the function of the resulting protein, if it is formed, and are thus rare in in vivo protein-coding sequences. Often such misformed proteins are targeted for proteolytic degradation. In addition, a frame shift mutation is very likely to cause a stop codon to be read, which truncates the creation of the protein.[9] One reason for the rareness of frame-shifted mutations' being inherited is that, if the protein being translated is essential for growth under the selective pressures the organism faces, absence of a functional protein may cause lethality before the organism is viable.[10]
Start/stop codons
Translation starts with a chain initiation codon (start codon). Unlike stop codons, the codon alone is not sufficient to begin the process. Nearby sequences (such as the Shine-Dalgarno sequence in E. Coli) and initiation factors are also required to start translation. The most common start codon is AUG which is read as methionine or, in bacteria, as formylmethionine. Alternative start codons (depending on the organism), include "GUG" or "UUG", which normally code for valine or leucine, respectively. However, when used as a start codon, these alternative start codons are translated as methionine or formylmethionine.[11]
The three stop codons have been given names: UAG is amber, UGA is opal (sometimes also called umber), and UAA is ochre. "Amber" was named by discoverers Richard Epstein and Charles Steinberg after their friend Harris Bernstein, whose last name means "amber" in German. The other two stop codons were named "ochre" and "opal" in order to keep the "color names" theme. Stop codons are also called "termination" or "nonsense" codons and they signal release of the nascent polypeptide from the ribosome due to binding of release factors in the absence of cognate tRNAs with anticodons complementary to these stop signals.[12]
Effect of mutations

Frameshift mutations altering the sequence reading frame, and nonsense mutations causing a stop codon are examples of point mutations. In addition, there may be missense mutations that cause exchange of one amino acid for another. Clinically important missense mutations generally change the properties of the coded amino acid residue between being basic, acidic polar or nonpolar, while nonsense mutations result in a stop codon.[8]: 266
Degeneracy of the genetic code
The genetic code has redundancy but no ambiguity (see the codon tables above for the full correlation). For example, although codons GAA and GAG both specify glutamic acid (redundancy), neither of them specifies any other amino acid (no ambiguity). The codons encoding one amino acid may differ in any of their three positions. For example the amino acid glutamic acid is specified by GAA and GAG codons (difference in the third position), the amino acid leucine is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position), while the amino acid serine is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second or third position).[6]: Chp 15
A position of a codon is said to be a fourfold degenerate site if any nucleotide at this position specifies the same amino acid. For example, the third position of the glycine codons (GGA, GGG, GGC, GGU) is a fourfold degenerate site, because all nucleotide substitutions at this site are synonymous; i.e., they do not change the amino acid. Only the third positions of some codons may be fourfold degenerate.[6]: Chp 15 A position of a codon is said to be a twofold degenerate site if only two of four possible nucleotides at this position specify the same amino acid. For example, the third position of the glutamic acid codons (GAA, GAG) is a twofold degenerate site. In twofold degenerate sites, the equivalent nucleotides are always either two purines (A/G) or two pyrimidines (C/U), so only transversional substitutions (purine to pyrimidine or pyrimidine to purine) in twofold degenerate sites are nonsynonymous.[6]: Chp 15 A position of a codon is said to be a non-degenerate site if any mutation at this position results in amino acid substitution. There is only one threefold degenerate site where changing three of the four nucleotides may have no effect on the amino acid (depending on what it is changed to), while changing the fourth possible nucleotide always results in an amino acid substitution. This is the third position of an isoleucine codon: AUU, AUC, or AUA all encode isoleucine, but AUG encodes methionine. In computation this position is often treated as a twofold degenerate site.[6]: Chp 15
There are three amino acids encoded by six different codons: serine, leucine, arginine. Only two amino acids are specified by a single codon; one of these is the amino-acid methionine, specified by the codon AUG, which also specifies the start of translation; the other is tryptophan, specified by the codon UGG. The degeneracy of the genetic code is what accounts for the existence of synonymous mutations.[6]: Chp 15
Degeneracy results because a triplet code designates 20 amino acids and a stop codon. Because there are four bases, triplet codons are required to produce at least 21 different codes. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required, then 4³ gives 64 possible codons, meaning that some degeneracy must exist.[6]: Chp 15
These properties of the genetic code make it more fault-tolerant for point mutations. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although codon usage bias restricts this in practice in many organisms; twofold degenerate codons can tolerate one out of the three possible point mutations at the third position. Since transition mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than transversion (purine to pyrimidine or vice-versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.[6]: Chp 15
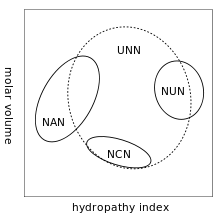
A practical consequence of redundancy is that some errors in the genetic code only cause a silent mutation or an error that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathy; NAN encodes average size hydrophilic residues.[14][15] These tendencies may result from that the aminoacyl tRNA synthetases related the such codons share a common ancestry.
Even so, single point mutations can still cause dysfunctional proteins. For example, a mutated hemoglobin gene causes sickle-cell disease. In the mutant hemoglobin a hydrophilic glutamate (Glu) is substituted by the hydrophobic valine (Val), that is, GAA or GAG becomes GUA or GUG. The substitution of glutamate by valine reduces the solubility of β-globin which causes hemoglobin to form linear polymers linked by the hydrophobic interaction between the valine groups causing sickle-cell deformation of erythrocytes. Sickle-cell disease is generally not caused by a de novo mutation. Rather it is selected for in malarial regions (in a way similar to thalassemia), as heterozygous people have some resistance to the malarial Plasmodium parasite (heterozygote advantage).[16]
These variable codes for amino acids are allowed because of modified bases in the first base of the anticodon of the tRNA, and the base-pair formed is called a wobble base pair. The modified bases include inosine and the Non-Watson-Crick U-G basepair.[17]
Variations to the standard genetic code
While slight variations on the standard code had been predicted earlier,[18] none were discovered until 1979, when researchers studying human mitochondrial genes discovered they used an alternative code. Many slight variants have been discovered since,[19] including various alternative mitochondrial codes,[20] as well as small variants such as Mycoplasma translating the codon UGA as tryptophan and Candida species translating CUG as a serine rather than a leucine.[21][22] In bacteria and archaea, GUG and UUG are common start codons. However, in rare cases, certain specific proteins may use alternative initiation (start) codons not normally used by that species.[23]
In certain proteins, non-standard amino acids are substituted for standard stop codons, depending upon associated signal sequences in the messenger RNA: UGA can code for selenocysteine and UAG can code for pyrrolysine as discussed in the relevant articles. Selenocysteine is now viewed as the 21st amino acid, and pyrrolysine is viewed as the 22nd. A detailed description of variations in the genetic code can be found at the NCBI web site.
Notwithstanding these differences, all known codes have strong similarities to each other, and the coding mechanism is the same for all organisms: three-base codons, tRNA, ribosomes, reading the code in the same direction and translating the code three letters at a time into sequences of amino acids.
Expanded genetic code
Since 2001, 40 non-natural amino acids have been added into protein by creating a unique codon (recoding) and a corresponding transfer-RNA:aminoacyl – tRNA-synthetase pair to encode it with diverse physicochemical and biological properties in order to be used as a tool to exploring protein structure and function or to create novel or enhanced proteins.[24]
Theories on the origin of the genetic code
Despite the variations that exist, the genetic codes used by all known forms of life are very similar. Since there are many possible genetic codes that are thought to have similar utility to the one used by Earth life, the theory of evolution suggests that the genetic code was established very early in the history of life. Phylogenetic analysis of transfer RNA suggests that tRNA molecules evolved before the present set of aminoacyl-tRNA synthetases.[25]
The genetic code is not a random assignment of codons to amino acids.[26] For example, amino acids that share the same biosynthetic pathway tend to have the same first base in their codons,[27] and amino acids with similar physical properties tend to have similar codons.[28][29]
There are four themes running through the many theories that seek to explain the evolution of the genetic code (and hence the origin of these patterns).[30]:
- Recent aptamer experiments show that some amino acids have a selective chemical affinity for the base triplets that code for them.[31] This suggests that the current complex translation mechanism involving tRNA and associated enzymes may be a later development, and that originally, protein sequences were directly templated on base sequences.
- That the standard modern genetic code grew from a simpler earlier code through a process of "biosynthetic expansion". Here the idea is that primordial life 'discovered' new amino acids (e.g., as by-products of metabolism) and later back-incorporated some of these into the machinery of genetic coding. Although much circumstantial evidence has been found to suggest that fewer different amino acids were used in the past than today.[32] precise and detailed hypotheses about exactly which amino acids entered the code in exactly what order has proved far more controversial.[33][34]
- That natural selection has led to codon assignments of the genetic code that minimize the effects of mutations.[35] A recent hypothesis [36] suggests that the triplet code was derived from codes that used longer than triplet codons. Longer than triplet decoding has higher degree of codon redundancy and is more error resistant than the triplet decoding. This feature could allow accurate decoding in the absence of highly complex translational machinery such as the ribosome.
- That viral agents that are competent in molecular syntax generate, arrange, rearrange and repair nucleotide sequences. According to the theory of biocommunication randomly derived mixtures of nucleotides (mutations) therefore doesn't play important roles in the evolution of the genetic code.[37]
See also
References
- ^ Turanov AA, Lobanov AV, Fomenko DE; et al. (2009). "Genetic code supports targeted insertion of two amino acids by one codon". Science. 323 (5911): 259–61. doi:10.1126/science.1164748. PMID 19131629.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Crick, Francis (1988). "Chapter 8: The genetic code". What mad pursuit: a personal view of scientific discovery. New York: Basic Books. pp. 89–101. ISBN 0-465-09138-5.
- ^ Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, Rottman F, O'Neal C (1965). "RNA codewords and protein synthesis, VII. On the general nature of the RNA code". Proc. Natl. Acad. Sci. U.S.A. 53 (5): 1161–8. PMC 301388. PMID 5330357.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "The Nobel Prize in Physiology or Medicine 1959" (Press release). The Royal Swedish Academy of Science. 1959. Retrieved 2010-02-27.
- ^ "The Nobel Prize in Physiology or Medicine 1968" (Press release). The Royal Swedish Academy of Science. 1968. Retrieved 2010-02-27.
- ^ a b c d e f g h i j k l Losick, Richard; Watson, James D.; Tania A. Baker; Bell, Stephen; Gann, Alexander; Levine, Michael W. (2008). Molecular Biology of the Gene. San Francisco: Pearson/Benjamin Cummings. ISBN 0-8053-9592-X.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Nakamoto T (2009). "Evolution and the universality of the mechanism of initiation of protein synthesis". Gene. 432 (1–2): 1–6. doi:10.1016/j.gene.2008.11.001. PMID 19056476.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Pamela K. Mulligan; King, Robert C.; Stansfield, William D. (2006). A dictionary of genetics. Oxford [Oxfordshire]: Oxford University Press. p. 608. ISBN 0-19-530761-5.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Isbrandt D, Hopwood JJ, von Figura K, Peters C (1996). "Two novel frameshift mutations causing premature stop codons in a patient with the severe form of Maroteaux-Lamy syndrome". Hum. Mutat. 7 (4): 361–3. doi:10.1002/(SICI)1098-1004(1996)7:4<361::AID-HUMU12>3.0.CO;2-0. PMID 8723688.
{{cite journal}}: Unknown parameter|doi_brokendate=ignored (|doi-broken-date=suggested) (help)CS1 maint: multiple names: authors list (link) - ^ Crow JF (1993). "How much do we know about spontaneous human mutation rates?". Environ. Mol. Mutagen. 21 (2): 122–9. PMID 8444142.
- ^ Touriol C, Bornes S, Bonnal S; et al. (2003). "Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons". Biology of the cell / under the auspices of the European Cell Biology Organization. 95 (3–4): 169–78. PMID 12867081.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ How nonsense mutations got their names
- ^ References for the image are found in Wikimedia Commons page at: Commons:File:Notable mutations.svg#References.
- ^ Yang; et al. (1990). Michel-Beyerle, M. E. (ed.). Reaction centers of photosynthetic bacteria: Feldafing-II-Meeting. Vol. 6. Berlin: Springer-Verlag. pp. 209–18. ISBN 3-540-53420-2.
{{cite book}}: Explicit use of et al. in:|author=(help) - ^ Füllen G, Youvan DC (1994). "Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering". Complexity International. 1.
- ^ Hebbel RP (2003). "Sickle hemoglobin instability: a mechanism for malarial protection". Redox Rep. 8 (5): 238–40. doi:10.1179/135100003225002826. PMID 14962356.
- ^ Varani G, McClain WH (2000). "The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems". EMBO Rep. 1 (1): 18–23. doi:10.1093/embo-reports/kvd001. PMC 1083677. PMID 11256617.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Crick FHC, Orgel LE (1973). "Directed panspermia". Icarus. 19: 341–6. doi:10.1016/0019-1035(73)90110-3.
It is a little surprising that organisms with somewhat different codes do not coexist.
(p. 344) (Further discussion) - ^ NCBI: "The Genetic Codes", Compiled by Andrzej (Anjay) Elzanowski and Jim Ostell
- ^ Jukes TH, Osawa S (1990). "The genetic code in mitochondria and chloroplasts". Experientia. 46 (11–12): 1117–26. doi:10.1007/BF01936921. PMID 2253709.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Santos, M.A.; Tuite, M.F. (1995). "The CUG codon is decoded in vivo as serine and not leucine in Candida albicans". Nucleic Acids Research. 23 (9): 1481–6. doi:10.1093/nar/23.9.1481. PMC 306886. PMID 7784200.
- ^ Butler, G.; Rasmussen, MD; Lin, MF; Santos, MA; Sakthikumar, S; Munro, CA; Rheinbay, E; Grabherr, M; Forche, A; et al. (2009). "Evolution of pathogenicity and sexual reproduction in eight Candida genomes". Nature. 459 (7247): 657–62. doi:10.1038/nature08064. PMID 19465905.
{{cite journal}}: Explicit use of et al. in:|first=(help) - ^ Genetic Code page in the NCBI Taxonomy section (Downloaded 27 April 2007.)
- ^ Xie J, Schultz PG. Adding amino acids to the genetic repertoire. Curr Opin Chem Biol. 2005 Dec;9(6):548-54. Epub 2005 Nov 2. Review. PMID: 16260173
Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009 Mar 27;16(3):323-36. Review. PMID: 19318213 - ^ De Pouplana, L.R. (1998). "Genetic code origins: tRNAs older than their synthetases?". Proceedings of the National Academy of Sciences. 95 (19): 11295. doi:10.1073/pnas.95.19.11295. PMC 21636. PMID 9736730.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Freeland SJ, Hurst LD (1998). "The genetic code is one in a million". J. Mol. Evol. 47 (3): 238–48. doi:10.1007/PL00006381. PMID 9732450.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Taylor FJ, Coates D (1989). "The code within the codons". BioSystems. 22 (3): 177–87. doi:10.1016/0303-2647(89)90059-2. PMID 2650752.
- ^ Di Giulio M (1989). "The extension reached by the minimization of the polarity distances during the evolution of the genetic code". J. Mol. Evol. 29 (4): 288–93. doi:10.1007/BF02103616. PMID 2514270.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Wong JT (1980). "Role of minimization of chemical distances between amino acids in the evolution of the genetic code". Proc. Natl. Acad. Sci. U.S.A. 77 (2): 1083–6. doi:10.1073/pnas.77.2.1083. PMC 348428. PMID 6928661.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Knight RD, Freeland SJ, Landweber LF (1999). "Selection, history and chemistry: the three faces of the genetic code". Trends Biochem. Sci. 24 (6): 241–7. doi:10.1016/S0968-0004(99)01392-4. PMID 10366854.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Knight RD, Landweber LF (1998). "Rhyme or reason: RNA-arginine interactions and the genetic code". Chem. Biol. 5 (9): R215–20. doi:10.1016/S1074-5521(98)90001-1. PMID 9751648.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Brooks DJ, Fresco JR, Lesk AM, Singh M (2002). "Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code". Mol. Biol. Evol. 19 (10): 1645–55. PMID 12270892.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Amirnovin R (1997). "An analysis of the metabolic theory of the origin of the genetic code". J. Mol. Evol. 44 (5): 473–6. doi:10.1007/PL00006170. PMID 9115171.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Ronneberg TA, Landweber LF, Freeland SJ (2000). "Testing a biosynthetic theory of the genetic code: fact or artifact?". Proc. Natl. Acad. Sci. U.S.A. 97 (25): 13690–5. doi:10.1073/pnas.250403097. PMC 17637. PMID 11087835.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Freeland SJ, Wu T, Keulmann N (2003). "The case for an error minimizing standard genetic code". Orig Life Evol Biosph. 33 (4–5): 457–77. doi:10.1023/A:1025771327614. PMID 14604186.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Baranov PV, Venin M, Provan G (2009). "Codon Size Reduction as the Origin of the Triplet Genetic Code". PLoS One. 4 (5): e5708. doi:10.1371/journal.pone.0005708. PMC 2682656. PMID 19479032.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Witzany, G (2009) Biocommunication and Natural Genome Editing. Springer, Netherlands.
Further reading
- Griffiths, Anthony J. F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (1999). An Introduction to genetic analysis (7th ed.). San Francisco: W.H. Freeman. ISBN 0-7167-3771-X.
{{cite book}}: CS1 maint: multiple names: authors list (link) - Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. ISBN 0-8153-3218-1.
{{cite book}}: CS1 maint: multiple names: authors list (link) - Lodish, Harvey F.; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James E. (2000). Molecular cell biology (4th ed.). San Francisco: W.H. Freeman. ISBN 0-7167-3706-X.
{{cite book}}: CS1 maint: multiple names: authors list (link)
