Betacoronavirus: Difference between revisions
Artoria2e5 (talk | contribs) mNo edit summary |
m +2019-nCov |
||
| Line 34: | Line 34: | ||
'''Betacoronaviruses''' are one of four genera of [[coronavirus]]es of the subfamily ''[[Orthocoronavirinae]]'' in the family ''[[Coronaviridae]]'', of the order ''[[Nidovirales]]''. They are [[Viral envelope|enveloped]], [[Sense (molecular biology)|positive-sense]], [[Base pair|single-stranded]] [[RNA virus]]es of [[Zoonosis|zoonotic origin]]. The coronavirus genera are each composed of varying viral lineages with the betacoronavirus genus containing four such lineages. In older literature, this genus is also known as '''group 2 conaviruses'''. |
'''Betacoronaviruses''' are one of four genera of [[coronavirus]]es of the subfamily ''[[Orthocoronavirinae]]'' in the family ''[[Coronaviridae]]'', of the order ''[[Nidovirales]]''. They are [[Viral envelope|enveloped]], [[Sense (molecular biology)|positive-sense]], [[Base pair|single-stranded]] [[RNA virus]]es of [[Zoonosis|zoonotic origin]]. The coronavirus genera are each composed of varying viral lineages with the betacoronavirus genus containing four such lineages. In older literature, this genus is also known as '''group 2 conaviruses'''. |
||
The Beta-CoVs of the greatest clinical importance concerning humans are [[Human coronavirus OC43|OC43]], and [[Human coronavirus HKU1|HKU1]] of the A lineage, [[Severe acute respiratory syndrome (SARS)|SARS-CoV]] of the B lineage, and [[Middle East respiratory syndrome coronavirus|MERS-CoV]] of the C lineage. MERS-CoV is the first betacoronavirus belonging to lineage C that is known to infect humans.<ref>ProMED. MERS-CoV–Eastern Mediterranean (06) (http://www.promedmail.org/)</ref><ref>{{cite journal|last1=Memish|first1=Z. A.|last2=Zumla|first2=A. I.|last3=Al-Hakeem|first3=R. F.|last4=Al-Rabeeah|first4=A. A.|last5=Stephens|first5=G. M.|year=2013|title=Family Cluster of Middle East Respiratory Syndrome Coronavirus Infections|journal=New England Journal of Medicine|volume=368|issue=26|pages=2487–94|doi=10.1056/NEJMoa1303729|pmc=|pmid=23718156|url=}}</ref> |
The Beta-CoVs of the greatest clinical importance concerning humans are [[Human coronavirus OC43|OC43]], and [[Human coronavirus HKU1|HKU1]] of the A lineage, [[Severe acute respiratory syndrome (SARS)|SARS-CoV]] and [[2019 novel coronavirus (2019-nCoV)|2019-nCov]] of the B lineage, <ref>{{cite web |title=Phylogeny of SARS-like betacoronaviruses |url=https://nextstrain.org/groups/blab/sars-like-cov |website=nextstrain |accessdate=18 January 2020}}</ref> and [[Middle East respiratory syndrome coronavirus|MERS-CoV]] of the C lineage. MERS-CoV is the first betacoronavirus belonging to lineage C that is known to infect humans.<ref>ProMED. MERS-CoV–Eastern Mediterranean (06) (http://www.promedmail.org/)</ref><ref>{{cite journal|last1=Memish|first1=Z. A.|last2=Zumla|first2=A. I.|last3=Al-Hakeem|first3=R. F.|last4=Al-Rabeeah|first4=A. A.|last5=Stephens|first5=G. M.|year=2013|title=Family Cluster of Middle East Respiratory Syndrome Coronavirus Infections|journal=New England Journal of Medicine|volume=368|issue=26|pages=2487–94|doi=10.1056/NEJMoa1303729|pmc=|pmid=23718156|url=}}</ref> |
||
The alpha- and beta-coronavirus genera descend from the bat gene pool.<ref name=b1>{{cite journal|last1=Woo|first1=P. C.|last2=Wang|first2=M.|last3=Lau|first3=S. K.|last4=Xu|first4=H.|last5=Poon|first5=R. W.|last6=Guo|first6=R.|last7=Wong|first7=B. H.|last8=Gao|first8=K.|last9=Tsoi|first9=H. W.|last10=Huang|first10=Y.|last11=Li|first11=K. S.|last12=Lam|first12=C. S.|last13=Chan|first13=K. H.|last14=Zheng|first14=B. J.|last15=Yuen|first15=K. Y.|year=2007|title=Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features|journal=Journal of Virology|volume=81|issue=4|pages=1574–85|doi=10.1128/JVI.02182-06|pmc=1797546|pmid=17121802|url=}}</ref><ref name=b2>{{cite journal|last1=Lau|first1=S. K.|last2=Woo|first2=P. C.|last3=Yip|first3=C. C.|last4=Fan|first4=R. Y.|last5=Huang|first5=Y.|last6=Wang|first6=M.|last7=Guo|first7=R.|last8=Lam|first8=C. S.|last9=Tsang|first9=A. K.|last10=Lai|first10=K. K.|last11=Chan|first11=K. H.|last12=Che|first12=X. Y.|last13=Zheng|first13=B. J.|last14=Yuen|first14=K. Y.|year=2012|title=Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits|journal=Journal of Virology|volume=86|issue=10|pages=5481–96|doi=10.1128/JVI.06927-11|pmc=3347282|pmid=22398294|url=}}</ref><ref name=b3>{{cite journal|last1=Lau|first1=S. K.|last2=Poon|first2=R. W.|last3=Wong|first3=B. H.|last4=Wang|first4=M.|last5=Huang|first5=Y.|last6=Xu|first6=H.|last7=Guo|first7=R.|last8=Li|first8=K. S.|last9=Gao|first9=K.|last10=Chan|first10=K. H.|last11=Zheng|first11=B. J.|last12=Woo|first12=P. C.|last13=Yuen|first13=K. Y.|year=2010|title=Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup|journal=Journal of Virology|volume=84|issue=21|pages=11385–94|doi=10.1128/JVI.01121-10|pmc=2953156|pmid=20702646|url=}}</ref> |
The alpha- and beta-coronavirus genera descend from the bat gene pool.<ref name=b1>{{cite journal|last1=Woo|first1=P. C.|last2=Wang|first2=M.|last3=Lau|first3=S. K.|last4=Xu|first4=H.|last5=Poon|first5=R. W.|last6=Guo|first6=R.|last7=Wong|first7=B. H.|last8=Gao|first8=K.|last9=Tsoi|first9=H. W.|last10=Huang|first10=Y.|last11=Li|first11=K. S.|last12=Lam|first12=C. S.|last13=Chan|first13=K. H.|last14=Zheng|first14=B. J.|last15=Yuen|first15=K. Y.|year=2007|title=Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features|journal=Journal of Virology|volume=81|issue=4|pages=1574–85|doi=10.1128/JVI.02182-06|pmc=1797546|pmid=17121802|url=}}</ref><ref name=b2>{{cite journal|last1=Lau|first1=S. K.|last2=Woo|first2=P. C.|last3=Yip|first3=C. C.|last4=Fan|first4=R. Y.|last5=Huang|first5=Y.|last6=Wang|first6=M.|last7=Guo|first7=R.|last8=Lam|first8=C. S.|last9=Tsang|first9=A. K.|last10=Lai|first10=K. K.|last11=Chan|first11=K. H.|last12=Che|first12=X. Y.|last13=Zheng|first13=B. J.|last14=Yuen|first14=K. Y.|year=2012|title=Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits|journal=Journal of Virology|volume=86|issue=10|pages=5481–96|doi=10.1128/JVI.06927-11|pmc=3347282|pmid=22398294|url=}}</ref><ref name=b3>{{cite journal|last1=Lau|first1=S. K.|last2=Poon|first2=R. W.|last3=Wong|first3=B. H.|last4=Wang|first4=M.|last5=Huang|first5=Y.|last6=Xu|first6=H.|last7=Guo|first7=R.|last8=Li|first8=K. S.|last9=Gao|first9=K.|last10=Chan|first10=K. H.|last11=Zheng|first11=B. J.|last12=Woo|first12=P. C.|last13=Yuen|first13=K. Y.|year=2010|title=Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup|journal=Journal of Virology|volume=84|issue=21|pages=11385–94|doi=10.1128/JVI.01121-10|pmc=2953156|pmid=20702646|url=}}</ref> |
||
Revision as of 10:14, 19 January 2020
| Betacoronavirus | |
|---|---|
| |
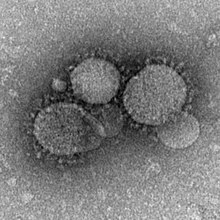
| MERS-CoV particles as seen by negative stain electron microscopy. Virions contain characteristic club-like projections emanating from the viral membrane. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Subfamily: | Orthocoronavirinae |
| Genus: | Betacoronavirus |
| Subgenera and Species[1] | |
| |
Betacoronaviruses are one of four genera of coronaviruses of the subfamily Orthocoronavirinae in the family Coronaviridae, of the order Nidovirales. They are enveloped, positive-sense, single-stranded RNA viruses of zoonotic origin. The coronavirus genera are each composed of varying viral lineages with the betacoronavirus genus containing four such lineages. In older literature, this genus is also known as group 2 conaviruses.
The Beta-CoVs of the greatest clinical importance concerning humans are OC43, and HKU1 of the A lineage, SARS-CoV and 2019-nCov of the B lineage, [2] and MERS-CoV of the C lineage. MERS-CoV is the first betacoronavirus belonging to lineage C that is known to infect humans.[3][4]
The alpha- and beta-coronavirus genera descend from the bat gene pool.[5][6][7]
Virology
Alpha- and betacoronaviruses mainly infect bats, but they also infect other species like humans, camels, and rabbits.[5][6][7][8] Beta-CoVs that have caused epidemics in humans generally induce fever and respiratory symptoms. They include:
- SARS-CoV, SARS disease.
- MERS-CoV, Middle East respiratory syndrome (MERS) disease.
- 2019-nCoV, 2019–20 outbreak of novel coronavirus (2019-nCoV).
Sequence
Coronaviruses have a large genome size that ranges from 26 to 32 kilobases.
As of May 2013, GenBank has 46 published complete genomes of the α-(group 1), β-(group 2), γ-(group 3), and δ-(group 4) CoVs.[9]
Classification

Within the genus Betacoronavirus (Group 2 CoV), four lineages (a, b, c, and d) are commonly recognized.
- Lineage A includes HCoV-OC43 and HCoV-HKU1 (various species)
- Lineage B includes SARS-CoV (various species) and 2019-nCoV
- Lineage C includes Tylonycteris bat coronavirus HKU4 (BtCoV-HKU4), Pipistrellus bat coronavirus HKU5 (BtCoV-HKU5), and MERS-CoV (various species)
- Lineage D includes Rousettus bat coronavirus HKU9 (BtCoV-HKU9)[10]
The four lineages are also named using greek letters or numerically.[9]
Morphology
The viruses of lineage A differ from all others in the genus in that they have a shorter spike-like protein called hemagglutinin esterase (HE).
The name Coronavirus is derived from the Latin “corona” meaning crown or halo, referring to their image under electron microscopy of crown-like spikes on their surface similar to the solar corona. This morphology is created by the viral spike (S) peplomers, which are proteins that populate the surface of the virus and determine host tropism. The order Nidovirales is named for the Latin nidus, which means nest. It refers to this order’s production of a 3' co-terminal nested set of subgenomic mRNA's during infection.[11]
See also
References
- ^ "Virus Taxonomy: 2018 Release" (html). International Committee on Taxonomy of Viruses (ICTV). October 2018. Retrieved 13 January 2019.
- ^ "Phylogeny of SARS-like betacoronaviruses". nextstrain. Retrieved 18 January 2020.
- ^ ProMED. MERS-CoV–Eastern Mediterranean (06) (http://www.promedmail.org/)
- ^ Memish, Z. A.; Zumla, A. I.; Al-Hakeem, R. F.; Al-Rabeeah, A. A.; Stephens, G. M. (2013). "Family Cluster of Middle East Respiratory Syndrome Coronavirus Infections". New England Journal of Medicine. 368 (26): 2487–94. doi:10.1056/NEJMoa1303729. PMID 23718156.
- ^ a b Woo, P. C.; Wang, M.; Lau, S. K.; Xu, H.; Poon, R. W.; Guo, R.; Wong, B. H.; Gao, K.; Tsoi, H. W.; Huang, Y.; Li, K. S.; Lam, C. S.; Chan, K. H.; Zheng, B. J.; Yuen, K. Y. (2007). "Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features". Journal of Virology. 81 (4): 1574–85. doi:10.1128/JVI.02182-06. PMC 1797546. PMID 17121802.
- ^ a b Lau, S. K.; Woo, P. C.; Yip, C. C.; Fan, R. Y.; Huang, Y.; Wang, M.; Guo, R.; Lam, C. S.; Tsang, A. K.; Lai, K. K.; Chan, K. H.; Che, X. Y.; Zheng, B. J.; Yuen, K. Y. (2012). "Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits". Journal of Virology. 86 (10): 5481–96. doi:10.1128/JVI.06927-11. PMC 3347282. PMID 22398294.
- ^ a b Lau, S. K.; Poon, R. W.; Wong, B. H.; Wang, M.; Huang, Y.; Xu, H.; Guo, R.; Li, K. S.; Gao, K.; Chan, K. H.; Zheng, B. J.; Woo, P. C.; Yuen, K. Y. (2010). "Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup". Journal of Virology. 84 (21): 11385–94. doi:10.1128/JVI.01121-10. PMC 2953156. PMID 20702646.
- ^ Zhang, Wei; Zheng, Xiao-Shuang; Agwanda, Bernard; Ommeh, Sheila; Zhao, Kai; Lichoti, Jacqueline; Wang, Ning; Chen, Jing; Li, Bei; Yang, Xing-Lou; Mani, Shailendra; Ngeiywa, Kisa-Juma; Zhu, Yan; Hu, Ben; Onyuok, Samson Omondi; Yan, Bing; Anderson, Danielle E.; Wang, Lin-Fa; Zhou, Peng; Shi, Zheng-Li (24 October 2019). "Serological evidence of MERS-CoV and HKU8-related CoV co-infection in Kenyan camels". Emerging Microbes & Infections. 8 (1): 1528–1534. doi:10.1080/22221751.2019.1679610.
- ^ a b Cotten, Matthew; Lam, Tommy T.; Watson, Simon J.; Palser, Anne L.; Petrova, Velislava; Grant, Paul; Pybus, Oliver G.; Rambaut, Andrew; Guan, Yi; Pillay, Deenan; Kellam, Paul; Nastouli, Eleni (2013-05-19). "Full-Genome Deep Sequencing and Phylogenetic Analysis of Novel Human Betacoronavirus - Vol. 19 No. 5 - May 2013 - CDC". Emerging Infectious Diseases. 19 (5): 736–42B. doi:10.3201/eid1905.130057. PMC 3647518. PMID 23693015. Retrieved 22 Apr 2014.
- ^ "ECDC Rapid Risk Assessment - Severe respiratory disease associated with a novel coronavirus" (PDF). 19 Feb 2013. Retrieved 22 Apr 2014.
- ^ Woo, P. C.; Huang, Y.; Lau, S. K.; Yuen, K. Y. (2010). "Coronavirus genomics and bioinformatics analysis". Viruses. 2 (8): 1804–20. doi:10.3390/v2081803. PMC 3185738. PMID 21994708.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
