DNA sequencing: Difference between revisions
| Line 140: | Line 140: | ||
This sequencing method and equipment were used to sequence the genome of the [[M13 bacteriophage]].<ref>{{cite journal | author = Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z | title = Single-molecule DNA sequencing of a viral genome. | journal = Science | volume = 320 | issue = 5872 | pages = 106–9 | date = 4 Apr 2008 | pmid = 18388294 | doi = 10.1126/science.1150427 | bibcode = 2008Sci...320..106H }}</ref> |
This sequencing method and equipment were used to sequence the genome of the [[M13 bacteriophage]].<ref>{{cite journal | author = Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z | title = Single-molecule DNA sequencing of a viral genome. | journal = Science | volume = 320 | issue = 5872 | pages = 106–9 | date = 4 Apr 2008 | pmid = 18388294 | doi = 10.1126/science.1150427 | bibcode = 2008Sci...320..106H }}</ref> |
||
probably obsolete: helicos is bankrupt |
|||
=== Single molecule real time (SMRT) sequencing === |
=== Single molecule real time (SMRT) sequencing === |
||
Revision as of 03:10, 3 January 2015
| Part of a series on |
| Genetics |
|---|
 |
DNA sequencing is the process of determining the precise order of nucleotides within a DNA molecule. It includes any method or technology that is used to determine the order of the four bases—adenine, guanine, cytosine, and thymine—in a strand of DNA. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.
Knowledge of DNA sequences has become indispensable for basic biological research, and in numerous applied fields such as diagnostic, biotechnology, forensic biology, virology and biological systematics. The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in the sequencing of complete DNA sequences, or genomes of numerous types and species of life, including the human genome and other complete DNA sequences of many animal, plant, and microbial species.
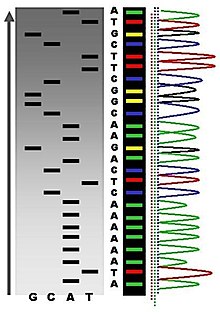
The first DNA sequences were obtained in the early 1970s by academic researchers using laborious methods based on two-dimensional chromatography. Following the development of fluorescence-based sequencing methods with automated analysis,[1] DNA sequencing has become easier and orders of magnitude faster.[2]
Use of sequencing
DNA sequencing may be used to determine the sequence of individual genes, larger genetic regions (i.e. clusters of genes or operons), full chromosomes or entire genomes. Sequencing provides the order of individual nucleotides in DNA or RNA (commonly represented as A, C, G, T, and U) isolated from cells of animals, plants, bacteria, archaea, or virtually any other source of genetic information. This is useful for:
- Molecular biology – studying the genome itself, how proteins are made, what proteins are made, identifying new genes and associations with diseases and phenotypes, and identifying potential drug targets
- Evolutionary biology – studying how different organisms are related and how they evolved
- Metagenomics – Identifying species present in a body of water, sewage, dirt, debris filtred from the air, or swab samples of organisms. Helpful in ecology, epidemiology, microbiome research, and other fields.
Less-precise information is produced by non-sequencing techniques like DNA fingerprinting. This information may be easier to obtain and is useful for:
- Detecting the presence of known genes for medical purposes (see genetic testing)
- Forensic identification
- Parental testing
The four canonical bases
The canonical structure of DNA has four bases: Thymine (T), Adenine (A), Cytosine (C), and Guanine (G). DNA sequencing is the determination of the physical order of these bases in a molecule of DNA. However, there are many other bases that may be present in a molecule. In some viruses (specifically, bacteriophage), cytosine is replaced by hydroxy methyl or hydroxy methyl glucose cytosine.[citation needed] In mammalian DNA, variant bases with methyl groups or phosphosulfate may be found.[citation needed] Depending on the sequencing technique, a particular modification may or may not be detected, eg, the 5mC (5 methyl cytosine) common in humans may or may not be detected.
History
Though the structure of DNA was established as a double helix in 1953,[3] several decades would pass before fragments of DNA could be reliably analyzed for their sequence in the laboratory. RNA sequencing was one of the earliest forms of nucleotide sequencing. The major landmark of RNA sequencing is the sequence of the first complete gene and the complete genome of Bacteriophage MS2, identified and published by Walter Fiers and his coworkers at the University of Ghent (Ghent, Belgium), in 1972[4] and 1976.[5]
The first method for determining DNA sequences involved a location-specific primer extension strategy established by Ray Wu at Cornell University in 1970.[6] DNA polymerase catalysis and specific nucleotide labeling, both of which figure prominently in current sequencing schemes, were used to sequence the cohesive ends of lambda phage DNA[7][8][9] Between 1970 and 1973, Wu, R Padmanabhan and colleagues demonstrated that this method can be employed to determine any DNA sequence using synthetic location-specific primers.[10][11][12][13] Frederick Sanger then adopted this primer-extension strategy to develop more rapid DNA sequencing methods at the MRC Centre, Cambridge, UK and published a method for "DNA sequencing with chain-terminating inhibitors" in 1977.[14] Walter Gilbert and Allan Maxam at Harvard also developed sequencing methods, including one for "DNA sequencing by chemical degradation".[15][16] In 1973, Gilbert and Maxam reported the sequence of 24 basepairs using a method known as wandering-spot analysis.[17] Advancements in sequencing were aided by the concurrent development of recombinant DNA technology, allowing DNA samples to be isolated from sources other than viruses.
The first full DNA genome to be sequenced was that of bacteriophage φX174 in 1977.[18] Medical Research Council scientists deciphered the complete DNA sequence of the Epstein-Barr virus in 1984, finding it to be 170 thousand base-pairs long.
A non-radioactive method for transferring the DNA molecules of sequencing reaction mixtures onto an immobilizing matrix during electrophoresis was developed by Pohl and co-workers in the early 80’s.[19][20] Followed by the commercialization of the DNA sequencer "Direct-Blotting-Electrophoresis-System GATC 1500" by GATC Biotech, which was intensively used in the framework of the EU genome-sequencing programme, the complete DNA sequence of the yeast Saccharomyces cerevisiae chromosome II.[21] Leroy E. Hood's laboratory at the California Institute of Technology announced the first semi-automated DNA sequencing machine in 1986.[22] This was followed by Applied Biosystems' marketing of the first fully automated sequencing machine, the ABI 370, in 1987 and by Dupont's Genesis 2000[23] which used a novel fluorescent labeling technique enabling all four dideoxynucleotides to be identified in a single lane. By 1990, the U.S. National Institutes of Health (NIH) had begun large-scale sequencing trials on Mycoplasma capricolum, Escherichia coli, Caenorhabditis elegans, and Saccharomyces cerevisiae at a cost of US$0.75 per base. Meanwhile, sequencing of human cDNA sequences called expressed sequence tags began in Craig Venter's lab, an attempt to capture the coding fraction of the human genome.[24] In 1995, Venter, Hamilton Smith, and colleagues at The Institute for Genomic Research (TIGR) published the first complete genome of a free-living organism, the bacterium Haemophilus influenzae. The circular chromosome contains 1,830,137 bases and its publication in the journal Science[25] marked the first published use of whole-genome shotgun sequencing, eliminating the need for initial mapping efforts. By 2001, shotgun sequencing methods had been used to produce a draft sequence of the human genome.[26][27]
Several new methods for DNA sequencing were developed in the mid to late 1990s. These techniques comprise the first of the "next-generation" sequencing methods. In 1996, Pål Nyrén and his student Mostafa Ronaghi at the Royal Institute of Technology in Stockholm published their method of pyrosequencing.[28] A year later, Pascal Mayer and Laurent Farinelli submitted patents to the World Intellectual Property Organization describing DNA colony sequencing.[29] Lynx Therapeutics published and marketed "Massively parallel signature sequencing", or MPSS, in 2000. This method incorporated a parallelized, adapter/ligation-mediated, bead-based sequencing technology and served as the first commercially available "next-generation" sequencing method, though no DNA sequencers were sold to independent laboratories.[30] In 2004, 454 Life Sciences marketed a parallelized version of pyrosequencing.[31][32] The first version of their machine reduced sequencing costs 6-fold compared to automated Sanger sequencing, and was the second of the new generation of sequencing technologies, after MPSS.[33]
The large quantities of data produced by DNA sequencing have also required development of new methods and programs for sequence analysis. Phil Green and Brent Ewing of the University of Washington described their phred quality score for sequencer data analysis in 1998.[34]
Basic methods
Maxam-Gilbert sequencing
Allan Maxam and Walter Gilbert published a DNA sequencing method in 1977 based on chemical modification of DNA and subsequent cleavage at specific bases.[15] Also known as chemical sequencing, this method allowed purified samples of double-stranded DNA to be used without further cloning. This method's use of radioactive labeling and its technical complexity discouraged extensive use after refinements in the Sanger methods had been made.
Maxam-Gilbert sequencing requires radioactive labeling at one 5' end of the DNA and purification of the DNA fragment to be sequenced. Chemical treatment then generates breaks at a small proportion of one or two of the four nucleotide bases in each of four reactions (G, A+G, C, C+T). The concentration of the modifying chemicals is controlled to introduce on average one modification per DNA molecule. Thus a series of labeled fragments is generated, from the radiolabeled end to the first "cut" site in each molecule. The fragments in the four reactions are electrophoresed side by side in denaturing acrylamide gels for size separation. To visualize the fragments, the gel is exposed to X-ray film for autoradiography, yielding a series of dark bands each corresponding to a radiolabeled DNA fragment, from which the sequence may be inferred.[15]
Chain-termination methods
The chain-termination method developed by Frederick Sanger and coworkers in 1977 soon became the method of choice, owing to its relative ease and reliability.[14][35] When invented, the chain-terminator method used fewer toxic chemicals and lower amounts of radioactivity than the Maxam and Gilbert method. Because of its comparative ease, the Sanger method was soon automated and was the method used in the first generation of DNA sequencers.
Sanger sequencing is the method which prevailed from the 80's until the mid-2000s. Over that period, great advances were made in the technique, such as fluorescent labelling, capillary electrophoresis, and general automation. These developments allowed much more efficient sequencing, leading to lower costs. The Sanger method, in mass production form, is the technology which produced the first human genome in 2001, ushering in the age of genomics. However, later in the decade, radically different approaches reached the market, bringing the cost per genome down from $100 million in 2001 to $10,000 in 2011.[36]
Advanced methods and de novo sequencing
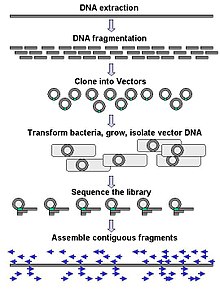
Large-scale sequencing often aims at sequencing very long DNA pieces, such as whole chromosomes, although large-scale sequencing can also be used to generate very large numbers of short sequences, such as found in phage display. For longer targets such as chromosomes, common approaches consist of cutting (with restriction enzymes) or shearing (with mechanical forces) large DNA fragments into shorter DNA fragments. The fragmented DNA may then be cloned into a DNA vector and amplified in a bacterial host such as Escherichia coli. Short DNA fragments purified from individual bacterial colonies are individually sequenced and assembled electronically into one long, contiguous sequence. Studies have shown that adding a size selection step to collect DNA fragments of uniform size can improve sequencing efficiency and accuracy of the genome assembly. In these studies, automated sizing has proven to be more reproducible and precise than manual gel sizing.[37][38][39]
The term "de novo sequencing" specifically refers to methods used to determine the sequence of DNA with no previously known sequence. De novo translates from Latin as "from the beginning". Gaps in the assembled sequence may be filled by primer walking. The different strategies have different tradeoffs in speed and accuracy; shotgun methods are often used for sequencing large genomes, but its assembly is complex and difficult, particularly with sequence repeats often causing gaps in genome assembly.
Most sequencing approaches use an in vitro cloning step to amplify individual DNA molecules, because their molecular detection methods are not sensitive enough for single molecule sequencing. Emulsion PCR[40] isolates individual DNA molecules along with primer-coated beads in aqueous droplets within an oil phase. A polymerase chain reaction (PCR) then coats each bead with clonal copies of the DNA molecule followed by immobilization for later sequencing. Emulsion PCR is used in the methods developed by Marguilis et al. (commercialized by 454 Life Sciences), Shendure and Porreca et al. (also known as "Polony sequencing") and SOLiD sequencing, (developed by Agencourt, later Applied Biosystems, now Life Technologies).[32][41][42]
Shotgun sequencing
Shotgun sequencing is a sequencing method designed for analysis of DNA sequences longer than 1000 base pairs, up to and including entire chromosomes. This method requires the target DNA to be broken into random fragments. After sequencing individual fragments, the sequences can be reassembled on the basis of their overlapping regions.[43]
Bridge PCR
Another method for in vitro clonal amplification is bridge PCR, in which fragments are amplified upon primers attached to a solid surface[29][44][45] and form "DNA colonies" or "DNA clusters". This method is used in the Illumina Genome Analyzer sequencers. Single-molecule methods, such as that developed by Stephen Quake's laboratory (later commercialized by Helicos) are an exception: they use bright fluorophores and laser excitation to detect base addition events from individual DNA molecules fixed to a surface, eliminating the need for molecular amplification.[46]
Next-generation methods

Next-generation sequencing applies to genome sequencing, genome resequencing, transcriptome profiling (RNA-Seq), DNA-protein interactions (ChIP-sequencing), and epigenome characterization.[47] Resequencing is necessary, because the genome of a single individual of a species will not indicate all of the genome variations among other individuals of the same species.
The high demand for low-cost sequencing has driven the development of high-throughput sequencing (or next-generation sequencing) technologies that parallelize the sequencing process, producing thousands or millions of sequences concurrently.[48][49] High-throughput sequencing technologies are intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods.[33] In ultra-high-throughput sequencing as many as 500,000 sequencing-by-synthesis operations may be run in parallel.[50][51][52]
| Method | Read length | Accuracy | Reads per run | Time per run | Cost per 1 million bases (in US$) | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| Single-molecule real-time sequencing (Pacific Bio) | 10,000 bp to 15,000 bp avg (14,000 bp N50); maximum read length >40,000 bases[55][56][57] | 99.9999% consensus accuracy; 87% single-read accuracy[58] | 50,000 per SMRT cell, or 500–1000 megabases[59][60] | 30 minutes to 4 hours[61] | $0.13–$0.60 | Longest read length. Fast. Detects 4mC, 5mC, 6mA.[62] | Moderate throughput. Equipment can be very expensive. |
| Ion semiconductor (Ion Torrent sequencing) | up to 400 bp | 98% | up to 80 million | 2 hours | $1 | Less expensive equipment. Fast. | Homopolymer errors. |
| Pyrosequencing (454) | 700 bp | 99.9% | 1 million | 24 hours | $10 | Long read size. Fast. | Runs are expensive. Homopolymer errors. |
| Sequencing by synthesis (Illumina) | 50 to 300 bp | 98% | up to 3 billion | 1 to 10 days, depending upon sequencer and specified read length[63] | $0.05 to $0.15 | Potential for high sequence yield, depending upon sequencer model and desired application. | Equipment can be very expensive. Requires high concentrations of DNA. |
| Sequencing by ligation (SOLiD sequencing) | 50+35 or 50+50 bp | 99.9% | 1.2 to 1.4 billion | 1 to 2 weeks | $0.13 | Low cost per base. | Slower than other methods. Have issue sequencing palindromic sequence.[64] |
| Chain termination (Sanger sequencing) | 400 to 900 bp | 99.9% | N/A | 20 minutes to 3 hours | $2400 | Long individual reads. Useful for many applications. | More expensive and impractical for larger sequencing projects. |
Massively parallel signature sequencing (MPSS)
The first of the next-generation sequencing technologies, massively parallel signature sequencing (or MPSS), was developed in the 1990s at Lynx Therapeutics, a company founded in 1992 by Sydney Brenner and Sam Eletr. MPSS was a bead-based method that used a complex approach of adapter ligation followed by adapter decoding, reading the sequence in increments of four nucleotides. This method made it susceptible to sequence-specific bias or loss of specific sequences. Because the technology was so complex, MPSS was only performed 'in-house' by Lynx Therapeutics and no DNA sequencing machines were sold to independent laboratories. Lynx Therapeutics merged with Solexa (later acquired by Illumina) in 2004, leading to the development of sequencing-by-synthesis, a simpler approach acquired from Manteia Predictive Medicine, which rendered MPSS obsolete. However, the essential properties of the MPSS output were typical of later "next-generation" data types, including hundreds of thousands of short DNA sequences. In the case of MPSS, these were typically used for sequencing cDNA for measurements of gene expression levels.[30]
Polony sequencing
The Polony sequencing method, developed in the laboratory of George M. Church at Harvard, was among the first next-generation sequencing systems and was used to sequence a full genome in 2005. It combined an in vitro paired-tag library with emulsion PCR, an automated microscope, and ligation-based sequencing chemistry to sequence an E. coli genome at an accuracy of >99.9999% and a cost approximately 1/9 that of Sanger sequencing.[65] The technology was licensed to Agencourt Biosciences, subsequently spun out into Agencourt Personal Genomics, and eventually incorporated into the Applied Biosystems SOLiD platform, which is now owned by Life Technologies, which was recently bought by Thermo Fisher Scientific.
454 pyrosequencing
A parallelized version of pyrosequencing was developed by 454 Life Sciences, which has since been acquired by Roche Diagnostics. The method amplifies DNA inside water droplets in an oil solution (emulsion PCR), with each droplet containing a single DNA template attached to a single primer-coated bead that then forms a clonal colony. The sequencing machine contains many picoliter-volume wells each containing a single bead and sequencing enzymes. Pyrosequencing uses luciferase to generate light for detection of the individual nucleotides added to the nascent DNA, and the combined data are used to generate sequence read-outs.[32] This technology provides intermediate read length and price per base compared to Sanger sequencing on one end and Solexa and SOLiD on the other.[33]
Illumina (Solexa) sequencing
Solexa, now part of Illumina, was founded by Shankar Balasubramanian and David Klenerman in 1998, and developed a sequencing method based on reversible dye-terminators technology, and engineered polymerases.[66] The terminated chemistry was developed internally at Solexa and the concept of the Solexa system was invented by Balasubramanian and Klenerman from Cambridge University's chemistry department. In 2004, Solexa acquired the company Manteia Predictive Medicine in order to gain a massivelly parallel sequencing technology based on "DNA Clusters", which involves the clonal amplification of DNA on a surface. The cluster technology was co-acquired with Lynx Therapeutics of California. Solexa Ltd. later merged with Lynx to form Solexa Inc.

In this method, DNA molecules and primers are first attached on a slide and amplified with polymerase so that local clonal DNA colonies, later coined "DNA clusters", are formed. To determine the sequence, four types of reversible terminator bases (RT-bases) are added and non-incorporated nucleotides are washed away. A camera takes images of the fluorescently labeled nucleotides, then the dye, along with the terminal 3' blocker, is chemically removed from the DNA, allowing for the next cycle to begin. Unlike pyrosequencing, the DNA chains are extended one nucleotide at a time and image acquisition can be performed at a delayed moment, allowing for very large arrays of DNA colonies to be captured by sequential images taken from a single camera.

Decoupling the enzymatic reaction and the image capture allows for optimal throughput and theoretically unlimited sequencing capacity. With an optimal configuration, the ultimately reachable instrument throughput is thus dictated solely by the analog-to-digital conversion rate of the camera, multiplied by the number of cameras and divided by the number of pixels per DNA colony required for visualizing them optimally (approximately 10 pixels/colony). In 2012, with cameras operating at more than 10 MHz A/D conversion rates and available optics, fluidics and enzymatics, throughput can be multiples of 1 million nucleotides/second, corresponding roughly to 1 human genome equivalent at 1x coverage per hour per instrument, and 1 human genome re-sequenced (at approx. 30x) per day per instrument (equipped with a single camera).[67]
SOLiD sequencing

Applied Biosystems' (now a Life Technologies brand) SOLiD technology employs sequencing by ligation. Here, a pool of all possible oligonucleotides of a fixed length are labeled according to the sequenced position. Oligonucleotides are annealed and ligated; the preferential ligation by DNA ligase for matching sequences results in a signal informative of the nucleotide at that position. Before sequencing, the DNA is amplified by emulsion PCR. The resulting beads, each containing single copies of the same DNA molecule, are deposited on a glass slide.[68] The result is sequences of quantities and lengths comparable to Illumina sequencing.[33] This sequencing by ligation method has been reported to have some issue sequencing palindromic sequences.[64]
Ion Torrent semiconductor sequencing
Ion Torrent Systems Inc. (now owned by Life Technologies) developed a system based on using standard sequencing chemistry, but with a novel, semiconductor based detection system. This method of sequencing is based on the detection of hydrogen ions that are released during the polymerisation of DNA, as opposed to the optical methods used in other sequencing systems. A microwell containing a template DNA strand to be sequenced is flooded with a single type of nucleotide. If the introduced nucleotide is complementary to the leading template nucleotide it is incorporated into the growing complementary strand. This causes the release of a hydrogen ion that triggers a hypersensitive ion sensor, which indicates that a reaction has occurred. If homopolymer repeats are present in the template sequence multiple nucleotides will be incorporated in a single cycle. This leads to a corresponding number of released hydrogens and a proportionally higher electronic signal.[69]

DNA nanoball sequencing
DNA nanoball sequencing is a type of high throughput sequencing technology used to determine the entire genomic sequence of an organism. The company Complete Genomics uses this technology to sequence samples submitted by independent researchers. The method uses rolling circle replication to amplify small fragments of genomic DNA into DNA nanoballs. Unchained sequencing by ligation is then used to determine the nucleotide sequence.[70] This method of DNA sequencing allows large numbers of DNA nanoballs to be sequenced per run and at low reagent costs compared to other next generation sequencing platforms.[71] However, only short sequences of DNA are determined from each DNA nanoball which makes mapping the short reads to a reference genome difficult.[70] This technology has been used for multiple genome sequencing projects and is scheduled to be used for more.[72]
Heliscope single molecule sequencing
Heliscope sequencing is a method of single-molecule sequencing developed by Helicos Biosciences. It uses DNA fragments with added poly-A tail adapters which are attached to the flow cell surface. The next steps involve extension-based sequencing with cyclic washes of the flow cell with fluorescently labeled nucleotides (one nucleotide type at a time, as with the Sanger method). The reads are performed by the Heliscope sequencer. The reads are short, up to 55 bases per run, but recent improvements allow for more accurate reads of stretches of one type of nucleotides.[73][74]
This sequencing method and equipment were used to sequence the genome of the M13 bacteriophage.[75]
probably obsolete: helicos is bankrupt
Single molecule real time (SMRT) sequencing
SMRT sequencing is based on the sequencing by synthesis approach. The DNA is synthesized in zero-mode wave-guides (ZMWs) – small well-like containers with the capturing tools located at the bottom of the well. The sequencing is performed with use of unmodified polymerase (attached to the ZMW bottom) and fluorescently labelled nucleotides flowing freely in the solution. The wells are constructed in a way that only the fluorescence occurring by the bottom of the well is detected. The fluorescent label is detached from the nucleotide upon its incorporation into the DNA strand, leaving an unmodified DNA strand. According to Pacific Biosciences, the SMRT technology developer, this methodology allows detection of nucleotide modifications (such as cytosine methylation). This happens through the observation of polymerase kinetics. This approach allows reads of 20,000 nucleotides or more, with average read lengths of 5 kilobases.[59][76]
Methods in development
DNA sequencing methods currently under development include labeling the DNA polymerase,[77] reading the sequence as a DNA strand transits through nanopores,[78][79] and microscopy-based techniques, such as atomic force microscopy or transmission electron microscopy that are used to identify the positions of individual nucleotides within long DNA fragments (>5,000 bp) by nucleotide labeling with heavier elements (e.g., halogens) for visual detection and recording.[80][81] Third generation technologies aim to increase throughput and decrease the time to result and cost by eliminating the need for excessive reagents and harnessing the processivity of DNA polymerase.[82]
Nanopore DNA sequencing
This method is based on the readout of electrical signals occurring at nucleotides passing by alpha-hemolysin pores covalently bound with cyclodextrin. The DNA passing through the nanopore changes its ion current. This change is dependent on the shape, size and length of the DNA sequence. Each type of the nucleotide blocks the ion flow through the pore for a different period of time. The method has a potential of development as it does not require modified nucleotides, however single nucleotide resolution is not yet available.[83]
Two main areas of nanopore sequencing in development are solid state nanopore sequencing, and protein based nanopore sequencing. Protein nanopore sequencing utilizes membrane protein complexes ∝-Hemolysin and MspA (Mycobacterium Smegmatis Porin A), which show great promise given their ability to distinguish between individual and groups of nucleotides.[84] Whereas, solid-state nanopore sequencing utilizes synthetic materials such as silicon nitride and aluminum oxide and it is preferred for its superior mechanical ability and thermal and chemical stability.[85] The fabrication method is essential for this type of sequencing given that the nanopore array can contain hundreds of pores with diameters smaller than eight nanometers.[84]
The concept originated from the idea that single stranded DNA or RNA molecules can be electrophoretically driven in a strict linear sequence through a biological pore that can be less than eight nanometers, and can be detected given that the molecules release an ionic current while moving through the pore. The pore contains a detection region capable of recognizing different bases, with each base generating various time specific signals corresponding to the sequence of bases as they cross the pore which are then evaluated.[85] When implementing this process it is important to note that precise control over the DNA transport through the pore is crucial for success. Various enzymes such as exonucleases and polymerases have been used to moderate this process by positioning them near the pore’s entrance.[86]
Tunnelling currents DNA sequencing
Another approach uses measurements of the electrical tunnelling currents across single-strand DNA as it moves through a channel. Depending on its electronic structure each base affects the tunnelling current differently, allowing differentiation between different bases.[87]
The use of tunnelling currents has the potential to sequence orders of magnitude faster than ionic current methods and the sequencing of several DNA oligomers and micro-RNA has already been achieved.[88]
Sequencing by hybridization
Sequencing by hybridization is a non-enzymatic method that uses a DNA microarray. A single pool of DNA whose sequence is to be determined is fluorescently labeled and hybridized to an array containing known sequences. Strong hybridization signals from a given spot on the array identifies its sequence in the DNA being sequenced.[89]
This method of sequencing utilizes binding characteristics of a library of short single stranded DNA molecules (oligonucleotides) also called DNA probes to reconstruct a target DNA sequence. Non-specific hybrids are removed by washing and the target DNA is eluted.[90] Hybrids are re-arranged such that the DNA sequence can be reconstructed. The benefit of this sequencing type is its ability to capture a large number of targets with a homogenous coverage.[91] Although a large number of chemicals and starting DNA is usually required. But, with the advent of solution-based hybridization much less equipment and chemicals are necessary.[90]
Sequencing with mass spectrometry
Mass spectrometry may be used to determine DNA sequences. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, or MALDI-TOF MS, has specifically been investigated as an alternative method to gel electrophoresis for visualizing DNA fragments. With this method, DNA fragments generated by chain-termination sequencing reactions are compared by mass rather than by size. The mass of each nucleotide is different from the others and this difference is detectable by mass spectrometry. Single-nucleotide mutations in a fragment can be more easily detected with MS than by gel electrophoresis alone. MALDI-TOF MS can more easily detect differences between RNA fragments, so researchers may indirectly sequence DNA with MS-based methods by converting it to RNA first.[92]
The higher resolution of DNA fragments permitted by MS-based methods is of special interest to researchers in forensic science, as they may wish to find single-nucleotide polymorphisms in human DNA samples to identify individuals. These samples may be highly degraded so forensic researchers often prefer mitochondrial DNA for its higher stability and applications for lineage studies. MS-based sequencing methods have been used to compare the sequences of human mitochondrial DNA from samples in a Federal Bureau of Investigation database[93] and from bones found in mass graves of World War I soldiers.[94]
Early chain-termination and TOF MS methods demonstrated read lengths of up to 100 base pairs.[95] Researchers have been unable to exceed this average read size; like chain-termination sequencing alone, MS-based DNA sequencing may not be suitable for large de novo sequencing projects. Even so, a recent study did use the short sequence reads and mass spectroscopy to compare single-nucleotide polymorphisms in pathogenic Streptococcus strains.[96]
Microfluidic Sanger sequencing
In microfluidic Sanger sequencing the entire thermocycling amplification of DNA fragments as well as their separation by electrophoresis is done on a single glass wafer (approximately 10 cm in diameter) thus reducing the reagent usage as well as cost.[97] In some instances researchers have shown that they can increase the throughput of conventional sequencing through the use of microchips.[98] Research will still need to be done in order to make this use of technology effective.
Microscopy-based techniques
This approach directly visualizes the sequence of DNA molecules using electron microscopy. The first identification of DNA base pairs within intact DNA molecules by enzymatically incorporating modified bases, which contain atoms of increased atomic number, direct visualization and identification of individually labeled bases within a synthetic 3,272 base-pair DNA molecule and a 7,249 base-pair viral genome has been demonstrated.[99]
RNAP sequencing
This method is based on use of RNA polymerase (RNAP), which is attached to a polystyrene bead. One end of DNA to be sequenced is attached to another bead, with both beads being placed in optical traps. RNAP motion during transcription brings the beads in closer and their relative distance changes, which can then be recorded at a single nucleotide resolution. The sequence is deduced based on the four readouts with lowered concentrations of each of the four nucleotide types, similarly to the Sanger method.[100]
RNA polymerase is attached to one end of a polystyrene bead and the other end is attached to the distal end of a DNA fragment. Each bead is then stuck in to an optical trap that levitates the beads. The interactions between the RNAP and the DNA result in a change in the length of the DNA between the two beads. This change is the measured with precision resulting in a single base resolution on a single DNA molecule. This is then repeated four times where each time there is a lower concentration of one of the four nucleotides, this shares some similarity with the primers used in the Sanger Sequencing method. A comparison is made between regions and sequence information is deduced by comparing the known sequence regions to the unknown sequence regions.[101]
In vitro virus high-throughput sequencing
A method has been developed to analyze full sets of protein interactions using a combination of 454 pyrosequencing and an in vitro virus mRNA display method. Specifically, this method covalently links proteins of interest to the mRNAs encoding them, then detects the mRNA pieces using reverse transcription PCRs. The mRNA may then be amplified and sequenced. The combined method was titled IVV-HiTSeq and can be performed under cell-free conditions, though its results may not be representative of in vivo conditions.[102]
Development initiatives

In October 2006, the X Prize Foundation established an initiative to promote the development of full genome sequencing technologies, called the Archon X Prize, intending to award $10 million to "the first Team that can build a device and use it to sequence 100 human genomes within 10 days or less, with an accuracy of no more than one error in every 100,000 bases sequenced, with sequences accurately covering at least 98% of the genome, and at a recurring cost of no more than $10,000 (US) per genome."[103]
Each year the National Human Genome Research Institute, or NHGRI, promotes grants for new research and developments in genomics. 2010 grants and 2011 candidates include continuing work in microfluidic, polony and base-heavy sequencing methodologies.[104]
Computational challenges
The sequencing technologies described here produce raw data that needs to be assembled into longer sequences such as complete genomes (sequence assembly). There are many computational challenges to achieve this, such as the evaluation of the raw sequence data which is done by programs and algorithms such as Phred and Phrap. Other challenges have to deal with repetitive sequences that often prevent complete genome assemblies because they occur in many places of the genome. As a consequence, many sequences may not be assigned to particular chromosomes. The production of raw sequence data is only the beginning of its detailed bioinformatical analysis.[105] Yet new methods for sequencing and correcting sequencing errors were developed.[106] [1]
Read Trimming
Sometimes, the raw reads produced by the sequencer are correct and precise only in a fraction of their length. Using the entire read may introduce artifacts in the downstream analyses like genome assembly, snp calling, or gene expression estimation. Two classes of trimming programs have been introduced, based on the window-based or the running-sum classes of algorithms.[107] This is a partial list of the trimming algorithms currently available, specifying the algorithm class they belong to:
- Cutadapt Running sum
- ConDeTri Window based
- ERNE-FILTER Running sum
- FASTX quality trimmer Window based
- PRINSEQ Window based
- Trimmomatic Window based
- SolexaQA Window based
- SolexaQA-BWA Running sum
- Sickle Window based
See also
|
References
- ^ Olsvik O, Wahlberg J, Petterson B, Uhlén M, Popovic T, Wachsmuth IK, Fields PI (January 1993). "Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains". J. Clin. Microbiol. 31 (1): 22–25. PMC 262614. PMID 7678018.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- ^ Pettersson E, Lundeberg J, Ahmadian A (February 2009). "Generations of sequencing technologies". Genomics. 93 (2): 105–11. doi:10.1016/j.ygeno.2008.10.003. PMID 18992322.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Watson JD, Crick FH (1953). "The structure of DNA". Cold Spring Harb. Symp. Quant. Biol. 18: 123–31. doi:10.1101/SQB.1953.018.01.020. PMID 13168976.
- ^ Min Jou W, Haegeman G, Ysebaert M, Fiers W (May 1972). "Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein". Nature. 237 (5350): 82–8. Bibcode:1972Natur.237...82J. doi:10.1038/237082a0. PMID 4555447.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M (April 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–7. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ . Cornell University http://mbg.cornell.edu/faculty-staff/faculty/wu.cfm.
{{cite web}}: Missing or empty|title=(help) - ^ PADMANABHAN, R; Ray Wu; Ernest Jay (June 1974). "Chemical Synthesis of a Primer and Its Use in the Sequence Analysis of the Lysozyme Gene of Bacteriophage T4". Proceedings of the National Academy of Sciences. 71 (6): 2510–2514. doi:10.1073/pnas.71.6.2510.
{{cite journal}}:|access-date=requires|url=(help) - ^ Onaga LA (June 2014). "Ray Wu as Fifth Business: Demonstrating Collective Memory in the History of DNA Sequencing". Studies in the History and Philosophy of Science. Part C. 46: 1–14. doi:10.1016/j.shpsc.2013.12.006. PMID 24565976. Retrieved 7 May 2014.
- ^ Wu, Ray (19 April 1972). "Nucleotide Sequence Analysis of DNA". Nature. 236 (68): 198–200. doi:10.1038/newbio236198a0. Retrieved 7 May 2014.
- ^ Padmanabhan, R; Ray Wu (1972). "Use of oligonucleotides of defined sequences as primers in DNA sequence analysis". Biochemical and Biophysical Research. 1295–1302. 48.
{{cite journal}}:|access-date=requires|url=(help) - ^ . Cornell http://mbg.cornell.edu/faculty-staff/faculty/wu.cfm. Retrieved 7 May 2014.
{{cite web}}: Missing or empty|title=(help) - ^ Wu, R; Padmanabhan (1973). "R". Biochemical and Biophysics Research. 55: 1092–1098.
{{cite journal}}:|access-date=requires|url=(help) - ^ Jay E, Bambara R, Padmanabhan R, Wu R (March 1974). "DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping". Nucleic Acids Research. 1: 331–353. doi:10.1093/nar/1.3.331. PMC 344020. PMID 10793670.
{{cite journal}}:|access-date=requires|url=(help)CS1 maint: multiple names: authors list (link) - ^ a b Sanger F, Nicklen S, Coulson AR (December 1977). "DNA sequencing with chain-terminating inhibitors". Proc. Natl. Acad. Sci. U.S.A. 74 (12): 5463–7. Bibcode:1977PNAS...74.5463S. doi:10.1073/pnas.74.12.5463. PMC 431765. PMID 271968.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Maxam AM, Gilbert W (February 1977). "A new method for sequencing DNA". Proc. Natl. Acad. Sci. U.S.A. 74 (2): 560–4. Bibcode:1977PNAS...74..560M. doi:10.1073/pnas.74.2.560. PMC 392330. PMID 265521.
- ^ Gilbert, W. DNA sequencing and gene structure. Nobel lecture, 8 December 1980.
- ^ Gilbert W, Maxam A (December 1973). "The Nucleotide Sequence of the lac Operator". Proc. Natl. Acad. Sci. U.S.A. 70 (12): 3581–4. Bibcode:1973PNAS...70.3581G. doi:10.1073/pnas.70.12.3581. PMC 427284. PMID 4587255.
- ^ Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (February 1977). "Nucleotide sequence of bacteriophage phi X174 DNA". Nature. 265 (5596): 687–95. Bibcode:1977Natur.265..687S. doi:10.1038/265687a0. PMID 870828.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Beck S, Pohl F M (1984). "DNA sequencing with direct blotting electrophoresis". EMBO J. 3 (12): 2905–2909. PMC 557787.
- ^ United States Patent 4,631,122 (1986)
- ^ Feldmann H; et al. (1994). "Complete DNA sequence of yeast chromosome II". EMBO J. 13 (24): 5795–5809. PMC 395553.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE (12 June 1986). "Fluorescence Detection in Automated DNA Sequence Analysis". Nature. 321 (6071): 674–79. doi:10.1038/321674a0. PMID 3713851.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K (16 October 1987). "A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides". Science. 238 (4825): 336–41. doi:10.1126/science.2443975. PMID 2443975.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF (June 1991). "Complementary DNA sequencing: expressed sequence tags and human genome project". Science. 252 (5013): 1651–6. Bibcode:1991Sci...252.1651A. doi:10.1126/science.2047873. PMID 2047873.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM (July 1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J; et al. (February 2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. doi:10.1038/35057062. PMID 11237011.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG; et al. (February 2001). "The sequence of the human genome". Science. 291 (5507): 1304–51. Bibcode:2001Sci...291.1304V. doi:10.1126/science.1058040. PMID 11181995.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P (1996). "Real-time DNA sequencing using detection of pyrophosphate release". Analytical Biochemistry. 242 (1): 84–9. doi:10.1006/abio.1996.0432. PMID 8923969.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Kawashima, Eric H.; Laurent Farinelli; Pascal Mayer (12 May 2005). "Patent: Method of nucleic acid amplification". Retrieved 22 December 2012{{inconsistent citations}}
{{cite web}}: CS1 maint: postscript (link) - ^ a b Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K (2000). "Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays". Nature Biotechnology. 18 (6). Nature Biotechnology: 630–634. doi:10.1038/76469. PMID 10835600.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stein RA (1 September 2008). "Next-Generation Sequencing Update". Genetic Engineering & Biotechnology News. 28 (15).
- ^ a b c Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA; et al. (September 2005). "Genome Sequencing in Open Microfabricated High Density Picoliter Reactors". Nature. 437 (7057): 376–80. Bibcode:2005Natur.437..376M. doi:10.1038/nature03959. PMC 1464427. PMID 16056220.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b c d Schuster SC (January 2008). "Next-generation sequencing transforms today's biology". Nat. Methods. 5 (1): 16–8. doi:10.1038/nmeth1156. PMID 18165802.
- ^ Ewing B, Green P (March 1998). "Base-calling of automated sequencer traces using phred. II. Error probabilities". Genome Res. 8 (3): 186–94. doi:10.1101/gr.8.3.186. PMID 9521922.
{{cite journal}}: Unknown parameter|doi_brokendate=ignored (|doi-broken-date=suggested) (help) - ^ Sanger F, Coulson AR (May 1975). "A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase". J. Mol. Biol. 94 (3): 441–8. doi:10.1016/0022-2836(75)90213-2. PMID 1100841.
- ^ Wetterstrand, Kris. "DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP)". National Human Genome Research Institute. Retrieved 30 May 2013.
- ^ Quail MA, Gu Y, Swerdlow H, Mayho M (2012). "Evaluation and optimisation of preparative semi-automated electrophoresis systems for Illumina library preparation". Electrophoresis. 33 (23): 3521–8. doi:10.1002/elps.201200128. PMID 23147856.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Duhaime MB, Deng L, Poulos BT, Sullivan MB (2012). "Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method". Environ. Microbiol. 14 (9): 2526–37. doi:10.1111/j.1462-2920.2012.02791.x. PMC 3466414. PMID 22713159.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012). "Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species". PLoS ONE. 7 (5): e37135. doi:10.1371/journal.pone.0037135. PMC 3365034. PMID 22675423.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Williams R, Peisajovich SG, Miller OJ, Magdassi S, Tawfik DS, Griffiths AD (2006). "Amplification of complex gene libraries by emulsion PCR". Nature methods. 3 (7): 545–550. doi:10.1038/nmeth896. PMID 16791213.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM (2005). "Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome". Science. 309 (5741): 1728–32. Bibcode:2005Sci...309.1728S. doi:10.1126/science.1117389. PMID 16081699.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Applied Biosystems' SOLiD technology
- ^ Staden R (11 June 1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Research. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874. PMID 461197.
- ^ P. Mayer,L. Farinelli, G. Matton, C. Adessi, G. Turcatti, J. J. Mermod, E. Kawashima.DNA colony massively parallel sequencing ams98 presentation
- ^ U.S. patent 5,641,658
- ^ Braslavsky I, Hebert B, Kartalov E, Quake SR (April 2003). "Sequence information can be obtained from single DNA molecules". Proc. Natl. Acad. Sci. U.S.A. 100 (7): 3960–4. Bibcode:2003PNAS..100.3960B. doi:10.1073/pnas.0230489100. PMC 153030. PMID 12651960.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ de Magalhães JP, Finch CE, Janssens G (2010). "Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions". Ageing Research Reviews. 9 (3): 315–323. doi:10.1016/j.arr.2009.10.006. PMC 2878865. PMID 19900591.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hall N (May 2007). "Advanced sequencing technologies and their wider impact in microbiology". J. Exp. Biol. 209 (Pt 9): 1518–1525. doi:10.1242/jeb.001370. PMID 17449817.

- ^ Church GM (January 2006). "Genomes for all". Sci. Am. 294 (1): 46–54. doi:10.1038/scientificamerican0106-46. PMID 16468433.(subscription required)
- ^ Kalb, Gilbert; Moxley, Robert (1992). Massively Parallel, Optical, and Neural Computing in the United States. IOS Press. ISBN 90-5199-097-9.[page needed]
- ^ ten Bosch JR, Grody WW (2008). "Keeping Up with the Next Generation". The Journal of Molecular Diagnostics. 10 (6): 484–492. doi:10.2353/jmoldx.2008.080027. PMC 2570630. PMID 18832462.

- ^ Tucker T, Marra M, Friedman JM (2009). "Massively Parallel Sequencing: The Next Big Thing in Genetic Medicine". The American Journal of Human Genetics. 85 (2): 142–154. doi:10.1016/j.ajhg.2009.06.022. PMC 2725244. PMID 19679224.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- ^ Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (1 January 2012). "A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers". BMC Genomics. 13 (1): 341. doi:10.1186/1471-2164-13-341. PMC 3431227. PMID 22827831.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
- ^ Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (1 January 2012). "Comparison of Next-Generation Sequencing Systems". Journal of Biomedicine and Biotechnology. 2012. Hindawi Publishing Corporation: 1–11. doi:10.1155/2012/251364. PMID 22829749.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
- ^ New Products: PacBio's RS II; Cufflinks | In Sequence | Sequencing | GenomeWeb
- ^ "After a Year of Testing, Two Early PacBio Customers Expect More Routine Use of RS Sequencer in 2012". GenomeWeb. 10 January 2012.(registration required)
- ^ Pacific Biosciences Introduces New Chemistry With Longer Read Lengths
- ^ http://www.nature.com/nmeth/journal/v10/n6/full/nmeth.2474.html
- ^ a b De novo bacterial genome assembly: a solved problem? | In between lines of code
- ^ Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Møller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK (25 August 2011). "Origins of the Strain Causing an Outbreak of Hemolytic–Uremic Syndrome in Germany". N Engl J Med. 365 (8): 709–717. doi:10.1056/NEJMoa1106920. PMID 21793740.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
- ^ Tran B, Brown AM, Bedard PL, Winquist E, Goss GD, Hotte SJ, Welch SA, Hirte HW, Zhang T, Stein LD, Ferretti V, Watt S, Jiao W, Ng K, Ghai S, Shaw P, Petrocelli T, Hudson TJ, Neel BG, Onetto N, Siu LL, McPherson JD, Kamel-Reid S, Dancey JE (1 January 2012). "Feasibility of real time next generation sequencing of cancer genes linked to drug response: Results from a clinical trial". Int. J. Cancer. 132 (7): 1547–1555. doi:10.1002/ijc.27817. PMID 22948899.
{{cite journal}}: CS1 maint: multiple names: authors list (link)(subscription required) - ^ Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ (2 October 2012). "The methylomes of six bacteria". Nucleic Acids Research. 40 (22): 11450–62. doi:10.1093/nar/gks891. PMC 3526280. PMID 23034806.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ van Vliet AH (1 January 2010). "Next generation sequencing of microbial transcriptomes: challenges and opportunities". FEMS Microbiology Letters. 302 (1): 1–7. doi:10.1111/j.1574-6968.2009.01767.x. PMID 19735299.

- ^ a b Huang YF, Chen SC, Chiang YS, Chen TH, Chiu KP (2012). "Palindromic sequence impedes sequencing-by-ligation mechanism". BMC systems biology. 6 Suppl 2: S10. doi:10.1186/1752-0509-6-S2-S10. PMID 23281822.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM (9 September 2005). "Accurate multiplex polony sequencing of an evolved bacterial genome". Science. 309 (5741): 1728–32. Bibcode:2005Sci...309.1728S. doi:10.1126/science.1117389. PMID 16081699.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG; et al. (2008). "Accurate whole human genome sequencing using reversible terminator chemistry". Nature. 456 (7218): 53–59. doi:10.1038/nature07517. PMC 2581791. PMID 18987734.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Mardis ER (2008). "Next-generation DNA sequencing methods". Annu Rev Genomics Hum Genet. 9: 387–402. doi:10.1146/annurev.genom.9.081307.164359. PMID 18576944.
- ^ Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM (July 2008). "A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning". Genome Res. 18 (7): 1051–63. doi:10.1101/gr.076463.108. PMC 2493394. PMID 18477713.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rusk N (2011). "Torrents of sequence". Nat Meth. 8 (1): 44–44. doi:10.1038/nmeth.f.330.
- ^ a b Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG; et al. (2010). "Human Genome Sequencing Using Unchained Base Reads in Self-Assembling DNA Nanoarrays". Science. 327 (5961): 78–81. Bibcode:2010Sci...327...78D. doi:10.1126/science.1181498. PMID 19892942.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Porreca GJ (2010). "Genome Sequencing on Nanoballs". Nature Biotechnology. 28 (1): 43–44. doi:10.1038/nbt0110-43. PMID 20062041.
- ^ Complete Genomics Press release, 2010
- ^ HeliScope Gene Sequencing / Genetic Analyzer System : Helicos BioSciences
- ^ Thompson JF, Steinmann KE (October 2010). "Single molecule sequencing with a HeliScope genetic analysis system". Current Protocols in Molecular Biology. Chapter 7: Unit7.10. doi:10.1002/0471142727.mb0710s92. PMC 2954431. PMID 20890904.
- ^ Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z (4 April 2008). "Single-molecule DNA sequencing of a viral genome". Science. 320 (5872): 106–9. Bibcode:2008Sci...320..106H. doi:10.1126/science.1150427. PMID 18388294.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ PacBio Sales Start to Pick Up as Company Delivers on Product Enhancements | In Sequence | Sequencing | GenomeWeb
- ^ "VisiGen Biotechnologies Inc. – Technology Overview". Visigenbio.com. Retrieved 15 November 2009.
- ^ "The Harvard Nanopore Group". Mcb.harvard.edu. Retrieved 15 November 2009.
- ^ "Nanopore Sequencing Could Slash DNA Analysis Costs".
- ^ US patent 20060029957, ZS Genetics, "Systems and methods of analyzing nucleic acid polymers and related components", issued 2005-07-14
- ^ Xu M, Fujita D, Hanagata N (December 2009). "Perspectives and challenges of emerging single-molecule DNA sequencing technologies". Small. 5 (23): 2638–49. doi:10.1002/smll.200900976. PMID 19904762.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schadt EE, Turner S, Kasarskis A (2010). "A window into third-generation sequencing". Human Molecular Genetics. 19 (R2): R227–40. doi:10.1093/hmg/ddq416. PMID 20858600.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H (12 May 2009). "Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore". Proceedings of the National Academy of Sciences of the United States of America. 106 (19): 7702–7. Bibcode:2009PNAS..106.7702S. doi:10.1073/pnas.0901054106. PMC 2683137. PMID 19380741.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b dela Torre R, Larkin J, Singer A, Meller A (2012). "Fabrication and characterization of solid-state nanopore arrays for high-throughput DNA sequencing". Nanotechnology. 23 (38): 385308. doi:10.1088/0957-4484/23/38/385308. PMC 3557807. PMID 22948520.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Pathak, B., Lofas, H., Prasongkit, J., Grigoriev, A., Ahuja, R., & Scheicher, R. H. (9 January 2012). Double-functionalized nanopore-embedded gold electrodes for rapid DNA sequencing. Applied Physics Letters, 100, 2.)
- ^ Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW (2008). "Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures". Proceedings of the National Academy of Sciences. 105 (4): 1176–1181. doi:10.1073/pnas.0710982105. PMC 2234111. PMID 18216253.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Massimiliano Di Ventra (2013). "Fast DNA sequencing by electrical means inches closer". Nanotechnology. 24 (34): 342501. doi:10.1088/0957-4484/24/34/342501.
- ^ Ohshiro T et al 2012 Sci. Rep. 2 501–7
- ^ Hanna GJ, Johnson VA, Kuritzkes DR, Richman DD, Martinez-Picado J, Sutton L, Hazelwood JD, D'Aquila RT (1 July 2000). "Comparison of Sequencing by Hybridization and Cycle Sequencing for Genotyping of Human Immunodeficiency Virus Type 1 Reverse Transcriptase". J. Clin. Microbiol. 38 (7): 2715–21. PMC 87006. PMID 10878069.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Morey M, Fernández-Marmiesse A, Castiñeiras D, Fraga JM, Couce ML, Cocho JA (2013). "A glimpse into past, present, and future DNA sequencing". Molecular Genetics and Metabolism. 110 (1–2): 3–24. doi:10.1016/j.ymgme.2013.04.024. PMID 23742747.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Qin Y, Schneider TM, Brenner MP (2012). Gibas, Cynthia (ed.). "Sequencing by Hybridization of Long Targets". PLoS ONE. 7 (5): e35819. doi:10.1371/journal.pone.0035819. PMC 3344849. PMID 22574124.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Edwards JR, Ruparel H, Ju J (2005). "Mass-spectrometry DNA sequencing". Mutation Research. 573 (1–2): 3–12. doi:10.1016/j.mrfmmm.2004.07.021. PMID 15829234.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hall TA, Budowle B, Jiang Y, Blyn L, Eshoo M, Sannes-Lowery KA, Sampath R, Drader JJ, Hannis JC, Harrell P, Samant V, White N, Ecker DJ, Hofstadler SA (2005). "Base composition analysis of human mitochondrial DNA using electrospray ionization mass spectrometry: A novel tool for the identification and differentiation of humans". Analytical Biochemistry. 344 (1): 53–69. doi:10.1016/j.ab.2005.05.028. PMID 16054106.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Howard R, Encheva V, Thomson J, Bache K, Chan YT, Cowen S, Debenham P, Dixon A, Krause JU, Krishan E, Moore D, Moore V, Ojo M, Rodrigues S, Stokes P, Walker J, Zimmermann W, Barallon R (15 June 2011). "Comparative analysis of human mitochondrial DNA from World War I bone samples by DNA sequencing and ESI-TOF mass spectrometry". Forensic science international. Genetics. 7 (1): 1–9. doi:10.1016/j.fsigen.2011.05.009. PMID 21683667.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Monforte JA, Becker CH (1 March 1997). "High-throughput DNA analysis by time-of-flight mass spectrometry". Nature Medicine. 3 (3): 360–362. doi:10.1038/nm0397-360. PMID 9055869.
- ^ Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM (8 February 2010). "Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics". Proceedings of the National Academy of Sciences. 107 (9): 4371–4376. Bibcode:2010PNAS..107.4371B. doi:10.1073/pnas.0911295107. PMID 20142485.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kan CW, Fredlake CP, Doherty EA, Barron AE (1 November 2004). "DNA sequencing and genotyping in miniaturized electrophoresis systems". Electrophoresis. 25 (21–22): 3564–3588. doi:10.1002/elps.200406161. PMID 15565709.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chen YJ, Roller EE, Huang X (2010). "DNA sequencing by denaturation: experimental proof of concept with an integrated fluidic device". Lab on Chip. 10 (9): 1153–1159. doi:10.1039/b921417h. PMID 20390134.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bell DC, Thomas WK, Murtagh KM, Dionne CA, Graham AC, Anderson JE, Glover WR (9 October 2012). "DNA Base Identification by Electron Microscopy". Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 18 (5): 1–5. Bibcode:2012MiMic..18.1049B. doi:10.1017/S1431927612012615. PMID 23046798.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pareek CS, Smoczynski R, Tretyn A (November 2011). "Sequencing technologies and genome sequencing". Journal of applied genetics. 52 (4): 413–35. doi:10.1007/s13353-011-0057-x. PMC 3189340. PMID 21698376.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pareek CS, Smoczynski R, Tretyn A (2011). "Sequencing technologies and genome sequencing". Journal of Applied Genetics. 52 (4): 413–435. doi:10.1007/s13353-011-0057-x. PMC 3189340. PMID 21698376.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fujimori S, Hirai N, Ohashi H, Masuoka K, Nishikimi A, Fukui Y, Washio T, Oshikubo T, Yamashita T, Miyamoto-Sato E (2012). "Next-generation sequencing coupled with a cell-free display technology for high-throughput production of reliable interactome data". Scientific reports. 2: 691. Bibcode:2012NatSR...2E.691F. doi:10.1038/srep00691. PMC 3466446. PMID 23056904.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "PRIZE Overview: Archon X PRIZE for Genomics"
- ^ The Future of DNA Sequencing
- ^ Jessica Severin, Marina Lizio, Jayson Harshbarger, Hideya Kawaji, Carsten O Daub, Yoshihide Hayashizaki, the FANTOM consortium, Nicolas Bertin, and Alistair RR Forrest. Interactive visualization and analysis of large-scale NGS data-sets using ZENBU. Nature Biotechnology, March 2014 doi:10.1038/nbt.2840
- ^ Shmilovici A. and Ben-Gal I. (2007). "Using a VOM Model for Reconstructing Potential Coding Regions in EST Sequences," (PDF). Journal of Computational Statistics, vol. 22, no. 1, 49 – 69.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM (2013). "An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis". PLoS ONE. 8 (12): e85024. doi:10.1371/journal.pone.0085024. PMC 3871669. PMID 24376861.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
