Capsid


A capsid is the protein shell of a virus, enclosing its genetic material. It consists of several oligomeric (repeating) structural subunits made of protein called protomers. The observable 3-dimensional morphological subunits, which may or may not correspond to individual proteins, are called capsomeres. The proteins making up the capsid are called capsid proteins or viral coat proteins (VCP). The capsid and inner genome is called the nucleocapsid.
Capsids are broadly classified according to their structure. The majority of the viruses have capsids with either helical or icosahedral[2][3] structure. Some viruses, such as bacteriophages, have developed more complicated structures due to constraints of elasticity and electrostatics.[4] The icosahedral shape, which has 20 equilateral triangular faces, approximates a sphere, while the helical shape resembles the shape of a spring, taking the space of a cylinder but not being a cylinder itself.[5] The capsid faces may consist of one or more proteins. For example, the foot-and-mouth disease virus capsid has faces consisting of three proteins named VP1–3.[6]
Some viruses are enveloped, meaning that the capsid is coated with a lipid membrane known as the viral envelope. The envelope is acquired by the capsid from an intracellular membrane in the virus' host; examples include the inner nuclear membrane, the Golgi membrane, and the cell's outer membrane.[7]
Once the virus has infected a cell and begins replicating itself, new capsid subunits are synthesized using the protein biosynthesis mechanism of the cell. In some viruses, including those with helical capsids and especially those with RNA genomes, the capsid proteins co-assemble with their genomes. In other viruses, especially more complex viruses with double-stranded DNA genomes, the capsid proteins assemble into empty precursor procapsids that include a specialized portal structure at one vertex. Through this portal, viral DNA is translocated into the capsid.[8]
Structural analyses of major capsid protein (MCP) architectures have been used to categorise viruses into lineages. For example, the bacteriophage PRD1, the algal virus Paramecium bursaria Chlorella virus-1 (PBCV-1), mimivirus and the mammalian adenovirus have been placed in the same lineage, whereas tailed, double-stranded DNA bacteriophages (Caudovirales) and herpesvirus belong to a second lineage.[9][10][11][12]
Specific shapes
Icosahedral


The icosahedral structure is extremely common among viruses. The icosahedron consists of 20 triangular faces delimited by 12 fivefold vertexes and consists of 60 asymmetric units. Thus, an icosahedral virus is made of 60N protein subunits. The number and arrangement of capsomeres in an icosahedral capsid can be classified using the "quasi-equivalence principle" proposed by Donald Caspar and Aaron Klug.[13] Like the Goldberg polyhedra, an icosahedral structure can be regarded as being constructed from pentamers and hexamers. The structures can be indexed by two integers h and k, with and ; the structure can be thought of as taking h steps from the edge of a pentamer, turning 60 degrees counterclockwise, then taking k steps to get to the next pentamer. The triangulation number T for the capsid is defined as:
In this scheme, icosahedral capsids contain 12 pentamers plus 10(T − 1) hexamers.[14][15] The T-number is representative of the size and complexity of the capsids.[16] Geometric examples for many values of h, k, and T can be found at List of geodesic polyhedra and Goldberg polyhedra.
Many exceptions to this rule exist: For example, the polyomaviruses and papillomaviruses have pentamers instead of hexamers in hexavalent positions on a quasi T = 7 lattice. Members of the double-stranded RNA virus lineage, including reovirus, rotavirus and bacteriophage φ6 have capsids built of 120 copies of capsid protein, corresponding to a T = 2 capsid, or arguably a T = 1 capsid with a dimer in the asymmetric unit. Similarly, many small viruses have a pseudo T = 3 (or P = 3) capsid, which is organized according to a T = 3 lattice, but with distinct polypeptides occupying the three quasi-equivalent positions [17]
T-numbers can be represented in different ways, for example T = 1 can only be represented as an icosahedron or a dodecahedron and, depending on the type of quasi-symmetry, T = 3 can be presented as a truncated dodecahedron, an icosidodecahedron, or a truncated icosahedron and their respective duals a triakis icosahedron, a rhombic triacontahedron, or a pentakis dodecahedron.[18][clarification needed]
Prolate
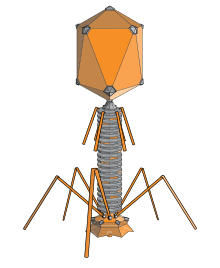
An elongated icosahedron is a common shape for the heads of bacteriophages. Such a structure is composed of a cylinder with a cap at either end. The cylinder is composed of 10 elongated triangular faces. The Q number (or Tmid), which can be any positive integer,[19] specifies the number of triangles, composed of asymmetric subunits, that make up the 10 triangles of the cylinder. The caps are classified by the T (or Tend) number.[20]
The bacterium E. coli is the host for bacteriophage T4 that has a prolate head structure. The bacteriophage encoded gp31 protein appears to be functionally homologous to E. coli chaperone protein GroES and able to substitute for it in the assembly of bacteriophage T4 virions during infection.[21] Like GroES, gp31 forms a stable complex with GroEL chaperonin that is absolutely necessary for the folding and assembly in vivo of the bacteriophage T4 major capsid protein gp23.[21]
Helical

Many rod-shaped and filamentous plant viruses have capsids with helical symmetry.[22] The helical structure can be described as a set of n 1-D molecular helices related by an n-fold axial symmetry.[23] The helical transformation are classified into two categories: one-dimensional and two-dimensional helical systems.[23] Creating an entire helical structure relies on a set of translational and rotational matrices which are coded in the protein data bank.[23] Helical symmetry is given by the formula P = μ x ρ, where μ is the number of structural units per turn of the helix, ρ is the axial rise per unit and P is the pitch of the helix. The structure is said to be open due to the characteristic that any volume can be enclosed by varying the length of the helix.[24] The most understood helical virus is the tobacco mosaic virus.[22] The virus is a single molecule of (+) strand RNA. Each coat protein on the interior of the helix bind three nucleotides of the RNA genome. Influenza A viruses differ by comprising multiple ribonucleoproteins, the viral NP protein organizes the RNA into a helical structure. The size is also different; the tobacco mosaic virus has a 16.33 protein subunits per helical turn,[22] while the influenza A virus has a 28 amino acid tail loop.[25]
Functions
The functions of the capsid are to:
- protect the genome,
- deliver the genome, and
- interact with the host.
The virus must assemble a stable, protective protein shell to protect the genome from lethal chemical and physical agents. These include extremes of pH or temperature and proteolytic and nucleolytic enzymes. For non-enveloped viruses, the capsid itself may be involved in interaction with receptors on the host cell, leading to penetration of the host cell membrane and internalization of the capsid. Delivery of the genome occurs by subsequent uncoating or disassembly of the capsid and release of the genome into the cytoplasm, or by ejection of the genome through a specialized portal structure directly into the host cell nucleus.
Origin and evolution
It has been suggested that many viral capsid proteins have evolved on multiple occasions from functionally diverse cellular proteins.[26] The recruitment of cellular proteins appears to have occurred at different stages of evolution so that some cellular proteins were captured and refunctionalized prior to the divergence of cellular organisms into the three contemporary domains of life, whereas others were hijacked relatively recently. As a result, some capsid proteins are widespread in viruses infecting distantly related organisms (e.g., capsid proteins with the jelly-roll fold), whereas others are restricted to a particular group of viruses (e.g., capsid proteins of alphaviruses).[26][27]
A computational model (2015) has shown that capsids may have originated before viruses and that they served as a means of horizontal transfer between replicator communities since these communities could not survive if the number of gene parasites increased, with certain genes being responsible for the formation of these structures and those that favored the survival of self-replicating communities.[28] The displacement of these ancestral genes between cellular organisms could favor the appearance of new viruses during evolution.[27]
See also
References
- ^ Asensio MA, Morella NM, Jakobson CM, Hartman EC, Glasgow JE, Sankaran B, et al. (September 2016). "A Selection for Assembly Reveals That a Single Amino Acid Mutant of the Bacteriophage MS2 Coat Protein Forms a Smaller Virus-like Particle". Nano Letters. 16 (9): 5944–50. Bibcode:2016NanoL..16.5944A. doi:10.1021/acs.nanolett.6b02948. OSTI 1532201. PMID 27549001. S2CID 16706951.
- ^ Lidmar J, Mirny L, Nelson DR (November 2003). "Virus shapes and buckling transitions in spherical shells". Physical Review E. 68 (5 Pt 1): 051910. arXiv:cond-mat/0306741. Bibcode:2003PhRvE..68e1910L. doi:10.1103/PhysRevE.68.051910. PMID 14682823. S2CID 6023873.
- ^ Vernizzi G, Olvera de la Cruz M (November 2007). "Faceting ionic shells into icosahedra via electrostatics". Proceedings of the National Academy of Sciences of the United States of America. 104 (47): 18382–6. Bibcode:2007PNAS..10418382V. doi:10.1073/pnas.0703431104. PMC 2141786. PMID 18003933.
- ^ Vernizzi G, Sknepnek R, Olvera de la Cruz M (March 2011). "Platonic and Archimedean geometries in multicomponent elastic membranes". Proceedings of the National Academy of Sciences of the United States of America. 108 (11): 4292–6. Bibcode:2011PNAS..108.4292V. doi:10.1073/pnas.1012872108. PMC 3060260. PMID 21368184.
- ^ Branden C, Tooze J (1991). Introduction to Protein Structure. New York: Garland. pp. 161–162. ISBN 978-0-8153-0270-4.
- ^ "Virus Structure (web-books.com)". Archived from the original on 2021-02-07. Retrieved 2007-07-10.
- ^ Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD (1994). Molecular Biology of the Cell (4th ed.). p. 280.
- ^ Newcomb WW, Homa FL, Brown JC (August 2005). "Involvement of the portal at an early step in herpes simplex virus capsid assembly". Journal of Virology. 79 (16): 10540–6. doi:10.1128/JVI.79.16.10540-10546.2005. PMC 1182615. PMID 16051846.
- ^ Krupovic M, Bamford DH (December 2008). "Virus evolution: how far does the double beta-barrel viral lineage extend?". Nature Reviews. Microbiology. 6 (12): 941–8. doi:10.1038/nrmicro2033. PMID 19008892. S2CID 31542714.
- ^ Forterre P (March 2006). "Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain". Proceedings of the National Academy of Sciences of the United States of America. 103 (10): 3669–74. Bibcode:2006PNAS..103.3669F. doi:10.1073/pnas.0510333103. PMC 1450140. PMID 16505372.
- ^ Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE (December 2005). "Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses". Proceedings of the National Academy of Sciences of the United States of America. 102 (52): 18944–9. doi:10.1073/pnas.0506383102. PMC 1323162. PMID 16357204.
- ^ Laurinmäki PA, Huiskonen JT, Bamford DH, Butcher SJ (December 2005). "Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35". Structure. 13 (12): 1819–28. doi:10.1016/j.str.2005.08.020. PMID 16338410.
- ^ Caspar DL, Klug A (1962). "Physical principles in the construction of regular viruses". Cold Spring Harbor Symposia on Quantitative Biology. 27: 1–24. doi:10.1101/sqb.1962.027.001.005. PMID 14019094.
- ^ Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, et al. (January 2009). "VIPERdb2: an enhanced and web API enabled relational database for structural virology". Nucleic Acids Research. 37 (Database issue): D436-42. doi:10.1093/nar/gkn840. PMC 2686430. PMID 18981051. Archived from the original on 2018-02-11. Retrieved 2011-03-18.
- ^ Johnson JE, Speir JA (2009). Desk Encyclopedia of General Virology. Boston: Academic Press. pp. 115–123. ISBN 978-0-12-375146-1.
- ^ Mannige RV, Brooks CL (March 2010). "Periodic table of virus capsids: implications for natural selection and design". PLOS ONE. 5 (3): e9423. Bibcode:2010PLoSO...5.9423M. doi:10.1371/journal.pone.0009423. PMC 2831995. PMID 20209096.
- ^ Sgro JY. "Virusworld". Institute for Molecular Virology. University of Wisconsin-Madison.
- ^ Damodaran KV, Reddy VS, Johnson JE, Brooks CL (December 2002). "A general method to quantify quasi-equivalence in icosahedral viruses". Journal of Molecular Biology. 324 (4): 723–37. doi:10.1016/S0022-2836(02)01138-5. PMID 12460573.
- ^ Luque A, Reguera D (June 2010). "The structure of elongated viral capsids". Biophysical Journal. 98 (12): 2993–3003. Bibcode:2010BpJ....98.2993L. doi:10.1016/j.bpj.2010.02.051. PMC 2884239. PMID 20550912.
- ^ Casjens S (2009). Desk Encyclopedia of General Virology. Boston: Academic Press. pp. 167–174. ISBN 978-0-12-375146-1.
- ^ a b Marusich EI, Kurochkina LP, Mesyanzhinov VV. Chaperones in bacteriophage T4 assembly. Biochemistry (Mosc). 1998;63(4):399-406
- ^ a b c Yamada S, Matsuzawa T, Yamada K, Yoshioka S, Ono S, Hishinuma T (December 1986). "Modified inversion recovery method for nuclear magnetic resonance imaging". The Science Reports of the Research Institutes, Tohoku University. Ser. C, Medicine. Tohoku Daigaku. 33 (1–4): 9–15. PMID 3629216.
- ^ a b c Aldrich RA (February 1987). "Children in cities--Seattle's KidsPlace program". Acta Paediatrica Japonica. 29 (1): 84–90. doi:10.1111/j.1442-200x.1987.tb00013.x. PMID 3144854. S2CID 33065417.
- ^ Racaniello VR, Enquist LW (2008). Principles of Virology, Vol. 1: Molecular Biology. Washington, D.C: ASM Press. ISBN 978-1-55581-479-3.
- ^ Ye Q, Guu TS, Mata DA, Kuo RL, Smith B, Krug RM, Tao YJ (26 December 2012). "Biochemical and structural evidence in support of a coherent model for the formation of the double-helical influenza A virus ribonucleoprotein". mBio. 4 (1): e00467–12. doi:10.1128/mBio.00467-12. PMC 3531806. PMID 23269829.
- ^ a b Krupovic M, Koonin EV (March 2017). "Multiple origins of viral capsid proteins from cellular ancestors". Proceedings of the National Academy of Sciences of the United States of America. 114 (12): E2401–E2410. Bibcode:2017PNAS..114E2401K. doi:10.1073/pnas.1621061114. PMC 5373398. PMID 28265094.
- ^ a b Krupovic M, Dolja VV, Koonin EV (July 2019). "Origin of viruses: primordial replicators recruiting capsids from hosts" (PDF). Nature Reviews. Microbiology. 17 (7): 449–458. doi:10.1038/s41579-019-0205-6. PMID 31142823. S2CID 169035711.
- ^ Jalasvuori M, Mattila S, Hoikkala V (2015). "Chasing the Origin of Viruses: Capsid-Forming Genes as a Life-Saving Preadaptation within a Community of Early Replicators". PLOS ONE. 10 (5): e0126094. Bibcode:2015PLoSO..1026094J. doi:10.1371/journal.pone.0126094. PMC 4425637. PMID 25955384.
Further reading
- Williams R (1 June 1979). The Geometrical Foundation of Natural Structure: A Source Book of Design. Dover Publications. pp. 142–144, Figures 4-49, 50, 51: Custers of 12 spheres, 42 spheres, 92 spheres. ISBN 978-0-486-23729-9.
- Pugh A (1 September 1976). Polyhedra: A Visual Approach. University of California Press. Chapter 6. The Geodesic Polyhedra of R. Buckminster Fuller and Related Polyhedra. ISBN 978-0-520-02926-2.
- Almansour I, Alhagri M, Alfares R, Alshehri M, Bakhashwain R, Maarouf A (January 2019). "IRAM: virus capsid database and analysis resource". Database: The Journal of Biological Databases and Curation. 2019. doi:10.1093/database/baz079. PMC 6637973. PMID 31318422.
External links
- IRAM-Virus Capsid Database and Analysis Resource Archived 2019-10-23 at the Wayback Machine




