Formate C-acetyltransferase
| formate C-acetyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.3.1.54 | ||||||||
| CAS no. | 9068-08-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
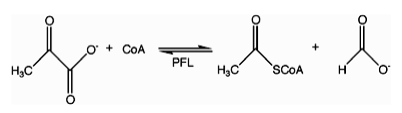
In enzymology, formate C-acetyltransferase (pyruvate formate lyase) (EC 2.3.1.54) is an enzyme. Pyruvate formate lyase is found in Escherichia coli[1] and other organisms. It helps regulate anaerobic glucose metabolism. Using radical non-redox chemistry, it catalyzes the reversible conversion of pyruvate and coenzyme-A into formate and acetyl-CoA. The reaction occurs as follows:
This enzyme belongs to the family of transferases, specifically those acyltransferases transferring groups other than aminoacyl groups. The systematic name of this enzyme class is acetyl-CoA:formate C-acetyltransferase. Other names in common use include pyruvate formate-lyase, pyruvic formate-lyase, and formate acetyltransferase. This enzyme participates in 3 metabolic pathways: pyruvate metabolism, propanoate metabolism, and butanoate metabolism.
Structural studies
As of late 2007, 8 structures have been solved for this class of enzymes, with PDB accession codes 1CM5, 1H16, 1H17, 1H18, 1MZO, 1QHM, 2PFL, and 3PFL.
Pyruvate formate lyase is a homodimer made of 85 kDa, 759-residue subunits. It has a 10-stranded beta/alpha barrel motif into which is inserted a beta finger that contains major catalytic residues. The active site of the enzyme, elucidated by x-ray crystallography, holds three essential amino acids that perform catalysis (Gly734, Cys418, and Cys419), three major residues that hold the substrate pyruvate close by (Arg435, Arg176, and Ala272), and two flanking hydrophobic residues (Trp333 and Phe432).[2]
Studies have found structural similarities between the active site of pyruvate formate lyase and that of Class I and Class III ribonucleotide reductase (RNR) enzymes.[2][3]
Mechanism
Roles of the three catalytic residues
- Gly734 (glycyl radical) – transfers the radical on and off Cys418, via Cys419
- Cys418 (thiyl radical) – does acylation chemistry on the carbon atom of the pyruvate carbonyl
- Cys419 (thiyl radical) – performs hydrogen-atom transfers
Steps
- The proposed mechanism for pyruvate formate lyase begins with radical transfer from Gly734 to Cys418, via Cys419.
- The Cys418 thiyl radical adds covalently to C2 (second carbon atom) of pyruvate, generating an acetyl-enzyme intermediate (which now contains the radical).
- The acetyl-enzyme intermediate releases a formyl radical that undergoes hydrogen-atom transfer with Cys419. This generates formate and a Cys419 radical.
- coenzyme-A comes in and undergoes hydrogen-atom transfer with the Cys419 radical to generate a coenzyme-A radical.
- The coenzyme-A radical then picks up the acetyl group from Cys418 to generate acetyl-CoA, leaving behind a Cys418 radical.
- Pyruvate formate lyase can then undergo radical transfer to put the radical back onto Gly734.
Note that each step is reversible.[4][5]
Regulation
Two additional enzymes regulate the “on” and “off” states of pyruvate formate lyase to regulate anaerobic glucose metabolism: pyruvate formate lyase activase (AE) and pyruvate formate lyase deactivase. Activated pyruvate formate lyase allows formation of acetyl-CoA, a small molecule important in the production of energy, when pyruvate is available. Deactivated pyruvate formate lyase, even with substrates present, does not catalyze the reaction.
Pyruvate formate lyase activase is part of the radical SAM (S-adenosylmethionine) superfamily. The enzyme turns pyruvate formate lyase “on” by converting Gly734 (G-H) into a Gly734 radical (G*) via a 5'-deoxyadenosyl radical (via a radical SAM).[6]
For more information about radical SAM activation and radical SAM enzymes, see the discussion by Wang et al., 2007.[7]
Pyruvate formate lyase deactivase turns pyruvate formate lyase “off” by quenching the Gly734 radical.[8] Furthermore, pyruvate formate lyase is sensitive to molecular oxygen (O2), the presence of which shuts the enzyme off.[9]
References
- ^ Knappe J, Blaschkowski HP, Grobner P, Schmitt T (1974). "Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate". Eur. J. Biochem. 50 (1): 253–63. doi:10.1111/j.1432-1033.1974.tb03894.x. PMID 4615902.
- ^ a b Becker A., Fritz-Wolf K., Kabsch W., Knappe J., Schultz S., Volker wagner A.F. Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. 1999 Nat. Struct. Biol. 6: 969–975.
- ^ Leppanen V.M., Merckel M.C., Ollis D.L., Wong K.K., Kozarich J.W., Goldman A. Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase. 1999 Structure 7: 733–744.
- ^ a b Becker, A., Kabsch W. X-ray structure of pyruvate formate-lyase in complex with pyruvate and CoA. How the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. 2002 J Biol Chem. 277(42): 40036–42.
- ^ a b Plaga, W., Wielhaber, G., Wallach, J., Knappe, J. Modification of Cys-418 of pyruvate formate-lyase by methacrylic acid, based on its radical mechanism. 2000 FEBS Lett. 466(1): 45–8.
- ^ Frey, M., Rothe, M., Wagner, AF., Knappe, J. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom. Studies of [2H]glycine-substituted enzyme and peptides homologous to the glycine 734 site. 1994 J Biol Chem. 269(17):12432–7.
- ^ Wang, SC., Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. 2007 Trends Biochem. Sci. 32(3): 101–10.
- ^ Nnyepi, MR., Peng, Y., Broderick, JB. Inactivation of E. coli pyruvate formate-lyase: role of AdhE and small molecules. 2007 Arch Biochem Biophys. 459(1):1–9.
- ^ Zhang, W., Wong, KK., Magliozzo, RS., Kozarich, JW. Inactivation of pyruvate formate-lyase by dioxygen: defining the mechanistic interplay of glycine 734 and cysteine 419 by rapid freeze-quench EPR. 2001 Biochemistry 40(13): 4123–30.