DNA computing
DNA computing is a form of computing which uses DNA, biochemistry and molecular biology, instead of the traditional silicon-based computer technologies. DNA computing, or, more generally, biomolecular computing, is a fast developing interdisciplinary area. Research and development in this area concerns theory, experiments and applications of DNA computing.
History
This field was initially developed by Leonard Adleman of the University of Southern California, in 1994.[1] Adleman demonstrated a proof-of-concept use of DNA as a form of computation which solved the seven-point Hamiltonian path problem. Since the initial Adleman experiments, advances have been made and various Turing machines have been proven to be constructible.[2] [3]
In 2002, researchers from the Weizmann Institute of Science in Rehovot, Israel, unveiled a programmable molecular computing machine composed of enzymes and DNA molecules instead of silicon microchips.[4] On April 28, 2004, Ehud Shapiro, Yaakov Benenson, Binyamin Gil, Uri Ben-Dor, and Rivka Adar at the Weizmann Institute announced in the journal Nature that they had constructed a DNA computer coupled with an input and output module which would theoretically be capable of diagnosing cancerous activity within a cell, and releasing an anti-cancer drug upon diagnosis. [5]
In 2009, biocomputing systems were coupled with standard silicon based chips for the first time. In this experiment, an enzyme based OR-Reset/AND-Reset logic system was achieved using field-effect Silicon chips. This advancement could yield great potential in the fields of Synthetic Biology, and Biomedical Engineering, as it marks the integration of biological and electro-mechanical systems on a sub-cellular level. [6]
Capabilities
DNA computing is fundamentally similar to parallel computing in that it takes advantage of the many different molecules of DNA to try many different possibilities at once.[7]
DNA computing also offers much lower power consumption than traditional silicon computers. DNA uses adenosine triphosphate (ATP) as fuel to allow ligation or as a means to heat the strand to cause disassociation.[8] Both strand hybridization and the hydrolysis of the DNA backbone can occur spontaneously, powered by the potential energy stored in DNA. Consumption of two ATP molecules releases 1.5 x 10−19 J. Even with a large number of transitions per second using two ATP molecules, power output is still low. For instance, Kahan reports 109 transitions per second with an energy consumption of 10−10 W,[9] and similarly Shapiro reports a system producing 7.5 x 1011 outputs in 4000 sec resulting in an energy consumption rate of ~ 10−10 W.[10]
For certain specialized problems, DNA computers are faster and smaller than any other computer built so far. Furthermore, particular mathematical computations have been demonstrated to work on a DNA computer. As an example, Aran Nayebi[11] has provided a general scalable implementation of Strassen's matrix multiplication algorithm on a DNA computer.
But DNA computing does not provide any new capabilities from the standpoint of computability theory, the study of which problems are computationally solvable using different models of computation. For example, if the space required for the solution of a problem grows exponentially with the size of the problem (EXPSPACE problems) on von Neumann machines, it still grows exponentially with the size of the problem on DNA machines. For very large EXPSPACE problems, the amount of DNA required is too large to be practical. (Quantum computing, on the other hand, does provide some interesting new capabilities.)
DNA computing overlaps with, but is distinct from, DNA nanotechnology. The latter uses the specificity of Watson-Crick basepairing and other DNA properties to make novel structures out of DNA. These structures can be used for DNA computing, but they do not have to be. Additionally, DNA computing can be done without using the types of molecules made possible by DNA nanotechnology.
Methods
There are multiple methods for building a computing device based on DNA, each with its own advantages and disadvantages. Most of these build the basic logic gates (AND, OR, NOT) associated with digital logic from a DNA basis. Some of the different bases include DNAzymes, deoxyoligonucleotides, enzymes, DNA tiling, and polymerase chain reaction.
DNAzymes
Catalytic DNA (deoxyribozyme or DNAzyme) catalyze a reaction when interacting with the appropriate input, such as a matching oligonucleotide. These DNAzymes are used to build logic gates analogous to digital logic in silicon; however, DNAzymes are limited to 1-, 2-, and 3-input gates with no current implementation for evaluating statements in series.
The DNAzyme logic gate changes its structure when it binds to a matching oligonucleotide and the fluorogenic substrate it is bonded to is cleaved free. While other materials can be used, most models use a fluorescence-based substrate because it is very easy to detect, even at the single molecule limit.[12] The amount of fluorescence can then be measured to tell whether or not a reaction took place. The DNAzyme that changes is then “used,” and cannot initiate any more reactions. Because of this, these reactions take place in a device such as a continuous stirred-tank reactor, where old product is removed and new molecules added.
Two commonly used DNAzymes are named E6 and 8-17. These are popular because they allow cleaving of a substrate in any arbitrary location.[13] Stojanovic and MacDonald have used the E6 DNAzymes to build the MAYA I[14] and MAYA II[15] machines, respectively; Stojanovic has also demonstrated logic gates using the 8-17 DNAzyme.[16] While these DNAzymes have been demonstrated to be useful for constructing logic gates, they are limited by the need for a metal cofactor to function, such as Zn2+ or Mn2+, and thus are not useful in vivo.[12][17]
A design called a stem loop, consisting of a single strand of DNA which has a loop at an end, are a dynamic structure that opens and closes when a piece of DNA bonds to the loop part. This effect has been exploited to create several logic gates. These logic gates have been used to create the computers MAYA I and MAYA II which can play tic-tac-toe to some extent.[18]
Enzymes
Enzyme based DNA computers are usually of the form of a simple Turing machine; there is analogous hardware, in the form of an enzyme, and software, in the form of DNA.[19]
Shapiro has demonstrated a DNA computer using the FokI enzyme[10] and expanded on his work by going on to show automata that diagnose and react to prostate cancer: under expression of the genes PPAP2B and GSTP1 and an over expression of PIM1 and HPN.[5] His automata evaluated the expression of each gene, one gene at a time, and on positive diagnosis then released a single strand DNA molecule (ssDNA) that is an antisense for MDM2. MDM2 is a repressor of protein 53, which itself is a tumor suppressor.[20] On negative diagnosis it was decided to release a suppressor of the positive diagnosis drug instead of doing nothing. A limitation of this implementation is that two separate automata are required, one to administer each drug. The entire process of evaluation until drug release took around an hour to complete. This method also requires transition molecules as well as the FokI enzyme to be present. The requirement for the FokI enzyme limits application in vivo, at least for use in “cells of higher organisms”.[9] It should also be pointed out that the 'software' molecules can be reused in this case.
Toehold exchange
DNA computers have also been constructed using the concept of toehold exchange. In this system, an input DNA strand binds to a sticky end, or toehold, on another DNA molecule, which allows it to displace another strand segment from the molecule. This allows the creation of modular logic components such as AND, OR, and NOT gates and signal amplifiers, which can be linked into arbitrarily large computers. This class of DNA computers does not require enzymes or any chemical capability of the DNA.[21]
Algorithmic self-assembly
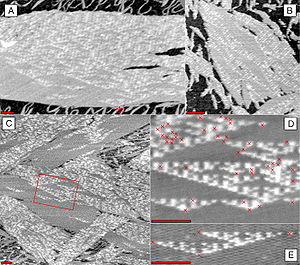
DNA nanotechnology has been applied to the related field of DNA computing. DNA tiles can be designed to contain multiple sticky ends with sequences chosen so that they act as Wang tiles. A DX array has been demonstrated whose assembly encodes an XOR operation; this allows the DNA array to implement a cellular automaton which generates a fractal called the Sierpinski gasket. This shows that computation can be incorporated into the assembly of DNA arrays, increasing its scope beyond simple periodic arrays.[22]
See also
- Biocomputers
- Computational gene
- Molecular electronics
- Peptide computing
- Parallel computing
- Quantum computing
- DNA code construction
References
- ^ Leonard M. Adleman (1994-11-11). "Molecular Computation Of Solutions To Combinatorial Problems" (PDF). Science. 266 (11): 1021–1024. doi:10.1126/science.7973651. — The first DNA computing paper. Describes a solution for the directed Hamiltonian path problem.
- ^ Dan Boneh, Christopher Dunworth, Richard J. Lipton, and Jiri Sgall (1996). "On the Computational Power of DNA" (PDF). DAMATH: Discrete Applied Mathematics and Combinatorial Operations Research and Computer Science. 71.
{{cite journal}}: CS1 maint: multiple names: authors list (link) — Describes a solution for the boolean satisfiability problem. - ^ Lila Kari, Greg Gloor, Sheng Yu (2000). "Using DNA to solve the Bounded Post Correspondence Problem". Theoretical Computer Science. 231 (2): 192–203.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) — Describes a solution for the bounded Post correspondence problem, a hard-on-average NP-complete problem. - ^ Lovgren, Stefan (2003-02-24). "Computer Made from DNA and Enzymes". National Geographic. Retrieved 2009-11-26.
- ^ a b
Yaakov Benenson1, Binyamin Gil, Uri Ben-Dor, Rivka Adar, Ehud Shapiro (2004-04-28). "An autonomous molecular computer for logical control of gene expression" (PDF). Nature. 429 (6990): 423–429. doi:10.1038/nature02551. PMID 15116117.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^
Melina Kramer, Marcos Pita, Jian Zhou, Maryna Ornatska, Arshak Poghossian, Michael Schoning, and Evgeny Katz (2008-12-03). "Coupling of Biocomputing Systems with Electronic Chips: Electronic Interface for Transduction of Biochemical Information". Journal of Physical Chemistry. 113: 2573–2579. doi:10.1021/jp808320s.
{{cite journal}}: Unknown parameter|DUPLICATE DATA: doi=ignored (help)CS1 maint: multiple names: authors list (link) - ^
David I. Lewin (2002). "DNA Computing". Computing in Science & Engineering. 4 (3): 5–8. doi:10.1109/5992.998634.
{{cite journal}}: Check date values in:|date=(help) - ^
C. H. Bennett (1973). "Logical Reversal of Computation". IBM Journal of Research and Development. 17 (6): 525–532. doi:10.1147/rd.176.0525.
{{cite journal}}: Check date values in:|date=(help) Link - ^ a b
Kahan, M; Gil, B; Adar, R; Shapiro, E (2008). "Towards Molecular Computers that Operate in a Biological Environment". Physica D: Nonlinear Phenomena. 237 (9): 1165–1172. doi:10.1016/j.physd.2008.01.027.
{{cite journal}}: Check date values in:|date=(help) - ^ a b
Benenson, Yaakov; Paz-Elizur, Tamar; Adar, Rivka; Keinan, Ehud; Livneh, Zvi; Shapiro, Ehud (2001). "Programmable and Autonomous Computing Machine Made of Biomolecules". Nature. 414 (6862): 430–434. doi:10.1038/35106533. PMID 11719800.
{{cite journal}}: Check date values in:|date=(help) - ^
Nayebi, A (2009). "Parallel DNA implementation of fast matrix multiplication techniques based on an n-moduli set". arXiv: 0912.0750: 1–15.
{{cite journal}}: Check date values in:|date=(help); External link in|journal= - ^ a b
Shimon Weiss (1999). "Fluorescence Spectroscopy of Single Biomolecules". Science. 283 (5408): 1676–1683. doi:10.1126/science.283.5408.1676.
{{cite journal}}: Check date values in:|date=(help) - ^
Stephen W. Santoro, Gerald F. Joyce (1997). "A General Purpose RNA-cleaving DNA Enzyme". Proc. Natl. Acad. Sci. 94 (9): 4262–4266. doi:10.1073/pnas.94.9.4262.
{{cite journal}}: Check date values in:|date=(help) - ^
Milan Stojanovic, Darko Stefanovic (2003). "A Deoxyribozyme-Based Molecular Automaton". Nature Biotechnology. 21 (9): 1069–1074. doi:10.1038/nbt862. PMID 12923549.
{{cite journal}}: Check date values in:|date=(help) - ^
MacDonald, Joanne; Li, Yang; Sutovic, Marko; Lederman, Harvey; Pendri, Kiran; Lu, Wanhong; Andrews, Benjamin L.; Stefanovic, Darko; Stojanovic, Milan N. (2006). "Medium Scale Integration of Molecular Logic Gates in an Automaton". Nano Letters. 6 (11): 2598–2603. doi:10.1021/nl0620684. PMID 17090098.
{{cite journal}}: Check date values in:|date=(help) - ^
Stojanovic, Milan N.; Mitchell, Tiffany Elizabeth; Stefanovic, Darko (2002). "Deoxyribozyme-Based Logic Gates". Journal of the American Chemical Society. 124: 3555–3561. doi:10.1021/ja016756v.
{{cite journal}}: Check date values in:|date=(help) - ^
Rani P. G. Cruz, Johanna B. Withers, Yingfu Li. (2004). "Dinucleotide Junction Cleavage: Versatility of 8-17 Deoxyribozyme". Chemistry & Biology. 11 (1): 57–67.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: multiple names: authors list (link) - ^ Darko Stefanovic's Group, Molecular Logic Gates and MAYA II, a second-generation tic-tac-toe playing automaton.
- ^ Shapiro, Ehud (1999-12-07). "A Mechanical Turing Machine: Blueprint for a Biomolecular Computer". Weizmann Institute of Science. Retrieved 2009-08-13.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^
Gareth L. Bond, Wenwei Hu, and Arnold J. Levine (2005). "MDM2 is a Central Node in the p53 Pathway: 12 Years and Counting". Current Cancer Drug Targets. 5 (1): 3–8. doi:10.2174/1568009053332627.
{{cite journal}}: Check date values in:|date=(help)CS1 maint: multiple names: authors list (link) - ^ Seelig, G.; Soloveichik, D.; Zhang, D. Y.; Winfree, E. (2006). "Enzyme-Free Nucleic Acid Logic Circuits". Science. 314 (5805): 1585. doi:10.1126/science.1132493. PMID 17158324.
- ^ a b Rothemund, Paul W. K. (2004). "Algorithmic Self-Assembly of DNA Sierpinski Triangles". PLoS Biology. 2 (12): 2041–2053. doi:10.1371/journal.pbio.0020424. ISSN 1544-9173. PMC 534809. PMID 15583715.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link)
Further reading
- Martyn Amos (2005). Theoretical and Experimental DNA Computation. Springer. ISBN 3-540-65773-8.
{{cite book}}: Unknown parameter|month=ignored (help) — The first general text to cover the whole field. - Gheorge Paun, Grzegorz Rozenberg, Arto Salomaa (1998). DNA Computing - New Computing Paradigms. Springer-Verlag. ISBN 3-540-64196-3.
{{cite book}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) — The book starts with an introduction to DNA-related matters, the basics of biochemistry and language and computation theory, and progresses to the advanced mathematical theory of DNA computing. - JB. Waldner (2007). Nanocomputers and Swarm Intelligence. ISTE. p. 189. ISBN 2746215160.
{{cite book}}: Unknown parameter|month=ignored (help) - Zoja Ignatova, Israel Martinez-Perez, Karl-Heinz Zimmermann (2008). DNA Computing Models. Springer. p. 288. ISBN 978-0-387-73635-8.
{{cite book}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) — A new general text to cover the whole field.
External links
- "DNA computing: the future for Basel?".
- How Stuff Works explanation
- Physics Web
- Ars Technica
- NY Times DNA Computer for detecting Cancer
- Bringing DNA computers to life, in Scientific American
- Japanese Researchers store information in bacteria DNA
- International Meeting on DNA Computing and Molecular Programming
- LiveScience.com-How DNA Could Power Computers
