MicroRNA: Difference between revisions
m →miRNA, DNA repair and cancer: Journal cites, added 2 DOIs using AWB (9488) |
Ccevol2013 (talk | contribs) |
||
| Line 85: | Line 85: | ||
==Evolution== |
==Evolution== |
||
MicroRNAs are significant phylogenetic markers because of their astonishingly low rate of evolution.<ref name=Wheeler2009/> Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs<ref name="Heimberg2008"/> and perhaps, ultimately, complex life.<ref name=Peterson2010>{{cite journal | author = Peterson KJ, Dietrich MR, McPeek MA | title = MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion | journal = BioEssays | volume = 31 | issue = 7 | pages = 736–47 | year = 2009 | month = July | pmid = 19472371 | doi = 10.1002/bies.200900033 | url = }}</ref> Indeed, rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.<ref name=Wheeler2009>{{cite journal | author = Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ | title = The deep evolution of metazoan microRNAs | journal = Evol. Dev. | volume = 11 | issue = 1 | pages = 50–68 | year = 2009 | pmid = 19196333 | doi = 10.1111/j.1525-142X.2008.00302.x | url = }}</ref><ref name=Heimberg2008>{{cite journal | author = Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ | title = MicroRNAs and the advent of vertebrate morphological complexity | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 105 | issue = 8 | pages = 2946–50 | year = 2008 | month = February | pmid = 18287013 | pmc = 2268565 | doi = 10.1073/pnas.0712259105 |bibcode = 2008PNAS..105.2946H }}</ref> |
MicroRNAs are significant phylogenetic markers because of their astonishingly low rate of evolution.<ref name=Wheeler2009/> MicroRNAs origin as a regulatory mechanism developed from previous RNAi machinery which was initially used as a defense against exogenous genetic material such as viruses. <ref>{{cite journal|last=Pashkovskiy|first=P. P.|coauthors=Ryazansky, S. S.|title=Biogenesis, evolution, and functions of plant microRNAs.|journal=Biochemistry-Moscow|year=2013|volume=78|pages=627-637|doi=10.1134/S0006297913060084|pmid=23980889|url=http://www.ncbi.nlm.nih.gov/pubmed/23980889}}</ref> Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs<ref name="Heimberg2008"/> and perhaps, ultimately, complex life.<ref name=Peterson2010>{{cite journal | author = Peterson KJ, Dietrich MR, McPeek MA | title = MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion | journal = BioEssays | volume = 31 | issue = 7 | pages = 736–47 | year = 2009 | month = July | pmid = 19472371 | doi = 10.1002/bies.200900033 | url = }}</ref> Indeed, rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.<ref name=Wheeler2009>{{cite journal | author = Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ | title = The deep evolution of metazoan microRNAs | journal = Evol. Dev. | volume = 11 | issue = 1 | pages = 50–68 | year = 2009 | pmid = 19196333 | doi = 10.1111/j.1525-142X.2008.00302.x | url = }}</ref><ref name=Heimberg2008>{{cite journal | author = Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ | title = MicroRNAs and the advent of vertebrate morphological complexity | journal = Proc. Natl. Acad. Sci. U.S.A. | volume = 105 | issue = 8 | pages = 2946–50 | year = 2008 | month = February | pmid = 18287013 | pmc = 2268565 | doi = 10.1073/pnas.0712259105 |bibcode = 2008PNAS..105.2946H }}</ref> |
||
New microRNAs are created in multiple different ways. Novel microRNAs can originate from the random formation of hairpins in "non-coding" sections of DNA (i.e. introns or intergene regions), but also by the duplication and modification of existing microRNAs.<ref name=Nozawa2010/>. MicroRNAs can also form from inverted duplications of protein-coding sequences, which allows for the creation of a foldback hairpin structure. <ref>{{cite journal|last=Allen|first=E.|coauthors=Z. X. Xie, A. M. Gustafson, G. H. Sung, J. W. Spatafora, and J. C. Carrington|title=Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana.|journal=Nature Genetics|year=2004|volume=36|pages=1282-1290|doi=10.1038/ng1478|url=http://www.nature.com/ng/journal/v36/n12/abs/ng1478.html}}</ref> The rate of evolution (i.e. nucleotide substitution) in recently originated microRNAs is comparable to that elsewhere in the non-coding DNA, implying evolution by neutral drift; however, older microRNAs have a much lower rate of change (often less than one substitution per hundred million years),<ref name=Peterson2010/> suggesting that once a microRNA gains a function it undergoes extreme purifying selection.<ref name=Nozawa2010/> Additionally, different regions within an miRNA gene seem to be under different evolutionary pressures, where regions that are vital for processing and function have much higher levels of conservation. <ref>{{cite journal|last=Warthmann|first=N.|coauthors=S. Das, C. Lanz, and D. Weigel|title=Comparative analysis of the MIR319a MicroRNA locus in Arabidopsis and related Brassicaceae.|journal=Molecular Biology and Evolution|year=2008|volume=25|issue=5|pages=892-902|doi=10.1093/molbev/msn029|pmid=18296705|url=http://www.ncbi.nlm.nih.gov/pubmed/18296705}}</ref> At this point, a microRNA is rarely lost from an animal's genome,<ref name=Peterson2010/> although microRNAs which are more recently derived (and thus presumably non-functional) are frequently lost.<ref name=Nozawa2010>{{cite journal | author = Nozawa M, Miura S, Nei M | title = Origins and evolution of microRNA genes in Drosophila species | journal = Genome Biol Evol | volume = 2 | issue = | pages = 180–9 | year = 2010 | pmid = 20624724 | pmc = 2942034 | doi = 10.1093/gbe/evq009 | url = }}</ref> In Arabidopsis thaliana, the net flux of miRNA genes has been predicted to be between 1.2 and 3.3 genes per million years. <ref>{{cite journal|last=Fahlgren|first=N.|coauthors=S. Jogdeo, K. D. Kasschau, C. M. Sullivan, E. J. Chapman, S. Laubinger, L. M. Smith, M. Dasenko, S. A. Givan, D. Weigel, and J. C. Carrington|title=MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana.|journal=Plant Cell|year=2010|volume=22|issue=4|pages=1074-1089|doi=10.1105/tpc.110.073999|url=http://www.plantcell.org/content/22/4/1074.abstract}}</ref> This makes them a valuable phylogenetic marker, and they are being looked upon as a possible solution to such outstanding phylogenetic problems as the relationships of arthropods.<ref name="pmid20486135">{{cite journal | author = Caravas J, Friedrich M | title = Of mites and millipedes: recent progress in resolving the base of the arthropod tree | journal = BioEssays | volume = 32 | issue = 6 | pages = 488–95 | year = 2010 | month = June | pmid = 20486135 | doi = 10.1002/bies.201000005 }}</ref> |
|||
MicroRNAs feature in the [[genome]]s of most eukaryotic organisms, from the brown algae<ref name="pmid20520714">{{cite journal | author = Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, ''et al''. | title = The Ectocarpus genome and the independent evolution of multicellularity in brown algae | journal = Nature | volume = 465 | issue = 7298 | pages = 617–21 | year = 2010 | month = June | pmid = 20520714 | doi = 10.1038/nature09016 |bibcode = 2010Natur.465..617C }}</ref> to the animals. Across all species, in excess of 5000 had been identified by March 2010.<ref name=Dimond2010>{{Cite news |
MicroRNAs feature in the [[genome]]s of most eukaryotic organisms, from the brown algae<ref name="pmid20520714">{{cite journal | author = Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, ''et al''. | title = The Ectocarpus genome and the independent evolution of multicellularity in brown algae | journal = Nature | volume = 465 | issue = 7298 | pages = 617–21 | year = 2010 | month = June | pmid = 20520714 | doi = 10.1038/nature09016 |bibcode = 2010Natur.465..617C }}</ref> to the animals. However, the difference in how these microRNAs function and the way they are processed suggests that microRNAs arose independently in plants and animals. <ref>{{cite journal|last=Cuperus|first=J. T.|coauthors=N. Fahlgren, and J. C. Carrington|title=Evolution and functional diversification of MIRNA genes.|journal=Plant Cell|year=2011|volume=23|issue=2|pages=431-442|doi=10.1105/tpc.110.082784|pmid=21317375|url=http://www.ncbi.nlm.nih.gov/pubmed/21317375}}</ref> Across all species, in excess of 5000 had been identified by March 2010.<ref name=Dimond2010>{{Cite news |
||
| author = Dimond PF |
| author = Dimond PF |
||
| date = 15 March 2010 | accessdate = 10 July 2010 |
| date = 15 March 2010 | accessdate = 10 July 2010 |
||
Revision as of 18:33, 16 November 2013
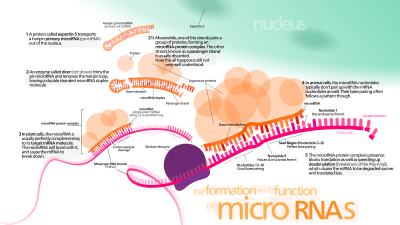

A microRNA (abbr. miRNA) is a small non-coding RNA molecule (ca. 22 nucleotides) found in plants and animals, which functions in transcriptional and post-transcriptional regulation of gene expression.[1] Encoded by eukaryotic nuclear DNA, miRNAs function via base-pairing with complementary sequences within mRNA molecules, usually resulting in gene silencing via translational repression or target degradation.[2][3] The human genome may encode over 1000 miRNAs,[4][5] which may target about 60% of mammalian genes[6][7] and are abundant in many human cell types.[8]
miRNAs are well conserved in eukaryotic organisms and are thought to be a vital and evolutionarily ancient component of genetic regulation.[9][10][11][12] While core components of the microRNA pathway are conserved between plants and animals, miRNA repertoires in the two kingdoms appear to have evolved independently with different modes of function.[13] Plant miRNAs usually have perfect or near-perfect pairing with their messenger RNA targets and induce gene repression through degradation of their target transcripts.[14][15] Plant miRNAs may bind their targets in both coding regions and untranslated regions.[15] In contrast, animal miRNAs typically exhibit only partial complementarity to their mRNA targets. A 'seed region' of about 6-8 nucleotides in length at the 5' end of an animal miRNA is thought to be an important determinant of target specificity.[6][16] Combinatorial regulation is a feature of miRNA regulation. A given miRNA may have multiple different mRNA targets, and a given target might similarly be targeted by multiple miRNAs.[17][18]
The first miRNAs were characterized in the early 1990s.[19] However, miRNAs were not recognized as a distinct class of biological regulators with conserved functions until the early 2000s. Since then, miRNA research has revealed multiple roles in negative regulation (transcript degradation and sequestering, translational suppression) and possible involvement in positive regulation (transcriptional and translational activation). By affecting gene regulation, miRNAs are likely to be involved in most biological processes.[20][21][22][23][24][25][26] Different sets of expressed miRNAs are found in different cell types and tissues.[27]
Aberrant expression of miRNAs has been implicated in numerous disease states, and miRNA-based therapies are under investigation.[28][29][30][31]
History
MicroRNAs were discovered in 1993 by Victor Ambros, Rosalind Lee and Rhonda Feinbaum during a study of the gene lin-14 in C. elegans development.[19] They found that LIN-14 protein abundance was regulated by a short RNA product encoded by the lin-4 gene. A 61-nucleotide precursor from the lin-4 gene matured to a 22-nucleotide RNA that contained sequences partially complementary to multiple sequences in the 3' UTR of the lin-14 mRNA. This complementarity was both necessary and sufficient to inhibit the translation of the lin-14 mRNA into the LIN-14 protein. Retrospectively, the lin-4 small RNA was the first microRNA to be identified, though at the time, it was thought to be a nematode idiosyncrasy. Only in 2000 was a second RNA characterized: let-7, which repressed lin-41, lin-14, lin-28, lin-42, and daf-12 expression during developmental stage transitions in C. elegans. let-7 was soon found to be conserved in many species,[32][33] indicating the existence of a wider phenomenon.
Nomenclature
Under a standard nomenclature system, names are assigned to experimentally confirmed miRNAs before publication of their discovery.[34][35] The prefix "mir" is followed by a dash and a number, the latter often indicating order of naming. For example, mir-123 was named and likely discovered prior to mir-456. The uncapitalized "mir-" refers to the pre-miRNA, while a capitalized "miR-" refers to the mature form. miRNAs with nearly identical sequences except for one or two nucleotides are annotated with an additional lower case letter. For example, miR-123a would be closely related to miR-123b. Pre-miRNAs that lead to 100% identical mature miRNAs but that are located at different places in the genome are indicated with an additional dash-number suffix. For example, the pre-miRNAs hsa-mir-194-1 and hsa-mir-194-2 lead to an identical mature miRNA (hsa-miR-194) but are located in different regions of the genome. Species of origin is designated with a three-letter prefix, e.g., hsa-miR-123 is a human (Homo sapiens) miRNA and oar-miR-123 is a sheep (Ovis aries) miRNA. Other common prefixes include 'v' for viral (miRNA encoded by a viral genome) and 'd' for Drosophila miRNA (a fruit fly commonly studied in genetic research). When two mature microRNAs originate from opposite arms of the same pre-miRNA, they are denoted with a -3p or -5p suffix. (In the past, this distinction was also made with 's' (sense) and 'as' (antisense)). When relative expression levels are known, an asterisk following the name indicates an miRNA expressed at low levels relative to the miRNA in the opposite arm of a hairpin. For example, miR-123 and miR-123* would share a pre-miRNA hairpin, but more miR-123 would be found in the cell.
Biogenesis

MicroRNAs are produced from either their own genes or from introns. A video of this process can be found here.
The majority of the characterized miRNA genes are intergenic or oriented antisense to neighboring genes and are therefore suspected to be transcribed as independent units.[36][36][37][38][39] However, in some cases a microRNA gene is transcribed together with its host gene; this provides a mean for coupled regulation of miRNA and protein-coding gene.[40] As much as 40% of miRNA genes may lie in the introns of protein and non-protein coding genes or even in exons of long nonprotein-coding transcripts.[41] These are usually, though not exclusively, found in a sense orientation,[42][43] and thus usually are regulated together with their host genes.[41][44][45] Other miRNA genes showing a common promoter include the 42-48% of all miRNAs originating from polycistronic units containing multiple discrete loops from which mature miRNAs are processed,[37][46] although this does not necessarily mean the mature miRNAs of a family will be homologous in structure and function. The promoters mentioned have been shown to have some similarities in their motifs to promoters of other genes transcribed by RNA polymerase II such as protein coding genes.[37][47] The DNA template is not the final word on mature miRNA production: 6% of human miRNAs show RNA editing (IsomiRs), the site-specific modification of RNA sequences to yield products different from those encoded by their DNA. This increases the diversity and scope of miRNA action beyond that implicated from the genome alone.
Transcription
miRNA genes are usually transcribed by RNA polymerase II (Pol II).[37][47] The polymerase often binds to a promoter found near the DNA sequence encoding what will become the hairpin loop of the pre-miRNA. The resulting transcript is capped with a specially modified nucleotide at the 5’ end, polyadenylated with multiple adenosines (a poly(A) tail),[37][42] and spliced. Animal miRNAs are initially transcribed as part of one arm of an ∼80 nucleotide RNA stem-loop that in turn forms part of a several hundred nucleotides long miRNA precursor termed a primary miRNA (pri-miRNA)s.[37][42] When a stem-loop precursor is found in the 3' UTR, a transcript may serve as a pri-miRNA and a mRNA.[42] RNA polymerase III (Pol III) transcribes some miRNAs, especially those with upstream Alu sequences, transfer RNAs (tRNAs), and mammalian wide interspersed repeat (MWIR) promoter units.[48]
Nuclear processing
A single pri-miRNA may contain from one to six miRNA precursors. These hairpin loop structures are composed of about 70 nucleotides each. Each hairpin is flanked by sequences necessary for efficient processing. The double-stranded RNA structure of the hairpins in a pri-miRNA is recognized by a nuclear protein known as DiGeorge Syndrome Critical Region 8 (DGCR8 or "Pasha" in invertebrates), named for its association with DiGeorge Syndrome. DGCR8 associates with the enzyme Drosha, a protein that cuts RNA, to form the "Microprocessor" complex.[49] In this complex, DGCR8 orients the catalytic RNase III domain of Drosha to liberate hairpins from pri-miRNAs by cleaving RNA about eleven nucleotides from the hairpin base (two helical RNA turns into the stem). The product resulting has a two-nucleotide overhang at its 3’ end; it has 3' hydroxyl and 5' phosphate groups. It is often termed as a pre-miRNA (precursor-miRNA).
Pre-miRNAs that are spliced directly out of introns, bypassing the Microprocessor complex, are known as "Mirtrons." Originally thought to exist only in Drosophila and C. elegans, mirtrons have now been found in mammals.[50]
Perhaps as many as 16% of pre-miRNAs may be altered through nuclear RNA editing.[51][52][53] Most commonly, enzymes known as adenosine deaminases acting on RNA (ADARs) catalyze adenosine to inosine (A to I) transitions. RNA editing can halt nuclear processing (for example, of pri-miR-142, leading to degradation by the ribonuclease Tudor-SN) and alter downstream processes including cytoplasmic miRNA processing and target specificity (e.g., by changing the seed region of miR-376 in the central nervous system).[51]
Nuclear export
Pre-miRNA hairpins are exported from the nucleus in a process involving the nucleocytoplasmic shuttle Exportin-5. This protein, a member of the karyopherin family, recognizes a two-nucleotide overhang left by the RNase III enzyme Drosha at the 3' end of the pre-miRNA hairpin. Exportin-5-mediated transport to the cytoplasm is energy-dependent, using GTP bound to the Ran protein.[54]
Cytoplasmic processing
In the cytoplasm, the pre-miRNA hairpin is cleaved by the RNase III enzyme Dicer.[55] This endoribonuclease interacts with the 3' end of the hairpin and cuts away the loop joining the 3' and 5' arms, yielding an imperfect miRNA:miRNA* duplex about 22 nucleotides in length.[55] Overall hairpin length and loop size influence the efficiency of Dicer processing, and the imperfect nature of the miRNA:miRNA* pairing also affects cleavage.[55][56] Although either strand of the duplex may potentially act as a functional miRNA, only one strand is usually incorporated into the RNA-induced silencing complex (RISC) where the miRNA and its mRNA target interact.
Biogenesis in plants
miRNA biogenesis in plants differs from animal biogenesis mainly in the steps of nuclear processing and export. Instead of being cleaved by two different enzymes, once inside and once outside the nucleus, both cleavages of the plant miRNA is performed by a Dicer homolog, called Dicer-like1 (DL1). DL1 is only expressed in the nucleus of plant cells, which indicates that both reactions take place inside the nucleus. Before plant miRNA:miRNA* duplexes are transported out of the nucleus, its 3' overhangs are methylated by a RNA methyltransferaseprotein called Hua-Enhancer1 (HEN1). The duplex is then transported out of the nucleus to the cytoplasm by a protein called Hasty (HST), an Exportin 5 homolog, where they disassemble and the mature miRNA is incorporated into the RISC.[57]
The RNA-induced silencing complex
The mature miRNA is part of an active RNA-induced silencing complex (RISC) containing Dicer and many associated proteins.[58] RISC is also known as a microRNA ribonucleoprotein complex (miRNP);[59] RISC with incorporated miRNA is sometimes referred to as "miRISC."
Dicer processing of the pre-miRNA is thought to be coupled with unwinding of the duplex. Generally, only one strand is incorporated into the miRISC, selected on the basis of its thermodynamic instability and weaker base-pairing relative to the other strand.[60][61][62] The position of the stem-loop may also influence strand choice.[63] The other strand, called the passenger strand due to its lower levels in the steady state, is denoted with an asterisk (*) and is normally degraded. In some cases, both strands of the duplex are viable and become functional miRNA that target different mRNA populations.[64]
Members of the Argonaute (Ago) protein family are central to RISC function. Argonautes are needed for miRNA-induced silencing and contain two conserved RNA binding domains: a PAZ domain that can bind the single stranded 3’ end of the mature miRNA and a PIWI domain that structurally resembles ribonuclease-H and functions to interact with the 5’ end of the guide strand. They bind the mature miRNA and orient it for interaction with a target mRNA. Some argonautes, for example human Ago2, cleave target transcripts directly; argonautes may also recruit additional proteins to achieve translational repression.[65] The human genome encodes eight argonaute proteins divided by sequence similarities into two families: AGO (with four members present in all mammalian cells and called E1F2C/hAgo in humans), and PIWI (found in the germ line and hematopoietic stem cells).[59][65]
Additional RISC components include TRBP [human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein],[66] PACT (protein activator of the interferon induced protein kinase (PACT), the SMN complex, fragile X mental retardation protein (FMRP), Tudor staphylococcal nuclease-domain-containing protein (Tudor-SN), the putative DNA helicase MOV10, and the RNA recognition motif containing protein TNRC6B.[54][67][68]
Mode of silencing
Gene silencing may occur either via mRNA degradation or preventing mRNA from being translated. For example, miR16 contains a sequence complementary to the AU-rich element found in the 3'UTR of many unstable mRNAs, such as TNF alpha or GM-CSF (ref Jing, Q., et al. Cell 2005 pmid = 15766526). It has been demonstrated that if there is complete complementation between the miRNA and target mRNA sequence, Ago2 can cleave the mRNA and lead to direct mRNA degradation. Yet, if there isn't complete complementation the silencing is achieved by preventing translation.[20]
miRNA turnover
Turnover of mature miRNA is needed for rapid changes in miRNA expression profiles. During miRNA maturation in the cytoplasm, uptake by the Argonaute protein is thought to stabilize the guide strand, while the opposite (* or "passenger") strand is preferentially destroyed. In what has been called a "Use it or lose it" strategy, Argonaute may preferentially retain miRNAs with many targets over miRNAs with few or no targets, leading to degradation of the non-targeting molecules.[69]
Decay of mature miRNAs in Caenorhabditis elegans is mediated by the 5´-to-3´ exoribonuclease XRN2, also known as Rat1p.[70] In plants, SDN (small RNA degrading nuclease) family members degrade miRNAs in the opposite (3'-to-5') direction. Similar enzymes are encoded in animal genomes, but their roles have not yet been described.[69]
Several miRNA modifications affect miRNA stability. As indicated by work in the model organism Arabidopsis thaliana (thale cress), mature plant miRNAs appear to be stabilized by the addition of methyl moieties at the 3' end. The 2'-O-conjugated methyl groups block the addition of uracil (U) residues by uridyltransferase enzymes, a modification that may be associated with miRNA degradation. However, uridylation may also protect some miRNAs; the consequences of this modification are incompletely understood. Uridylation of some animal miRNAs has also been reported. Both plant and animal miRNAs may be altered by addition of adenine (A) residues to the 3' end of the miRNA. An extra A added to the end of mammalian miR-122, a liver-enriched miRNA important in Hepatitis C, stabilizes the molecule, and plant miRNAs ending with an adenine residue have slower decay rates.[69]
Cellular functions

The function of miRNAs appears to be in gene regulation. For that purpose, a miRNA is complementary to a part of one or more messenger RNAs (mRNAs). Animal miRNAs are usually complementary to a site in the 3' UTR whereas plant miRNAs are usually complementary to coding regions of mRNAs.[72] Perfect or near perfect base pairing with the target RNA promotes cleavage of the RNA.[73] This is the primary mode of plant miRNAs.[74] In animals miRNAs more often have only partly the right sequence of nucleotides to bond with the target mRNA. The match-ups are imperfect. For partially complementary microRNAs to recognise their targets, nucleotides 2–7 of the miRNA (its 'seed region'[6][16]) still have to be perfectly complementary.[75] Animal miRNAs inhibit protein translation of the target mRNA[76] (this exists in plants as well but is less common).[74] MicroRNAs that are partially complementary to a target can also speed up deadenylation, causing mRNAs to be degraded sooner.[77] While degradation of miRNA-targeted mRNA is well documented, whether or not translational repression is accomplished through mRNA degradation, translational inhibition, or a combination of the two is hotly debated. Recent work on miR-430 shows that translational repression is caused by the disruption of translation initiation, independent of mRNA deadenylation.[78]
miRNAs occasionally also cause histone modification and DNA methylation of promoter sites, which affects the expression of target genes.[79][80]
Nine mechanisms of miRNA action are described and assembled in a unified mathematical model:[71]
- Cap-40S initiation inhibition;
- 60S Ribosomal unit joining inhibition;
- Elongation inhibition;
- Ribosome drop-off (premature termination);
- Co-translational nascent protein degradation;
- Sequestration in P-bodies;
- mRNA Decay (destabilisation);
- mRNA Cleavage;
- Transcriptional inhibition through microRNA-mediated chromatin reorganization following by gene silencing.
It is often impossible to discern these mechanisms using the experimental data about stationary reaction rates. Nevertheless, they are differentiated in dynamics and have different kinetic signatures.[71]
Unlike plant microRNAs, the animal microRNAs target a diverse set of genes.[16] However, genes involved in functions common to all cells, such as gene expression, have relatively fewer microRNA target sites and seem to be under selection to avoid targeting by microRNAs.[81]
dsRNA can also activate gene expression, a mechanism that has been termed "small RNA-induced gene activation" or RNAa. dsRNAs targeting gene promoters can induce potent transcriptional activation of associated genes. This was demonstrated in human cells using synthetic dsRNAs termed small activating RNAs (saRNAs),[82] but has also been demonstrated for endogenous microRNA.[83]
Interactions between microRNAs and complementary sequences on genes and even pseudogenes that share sequence homology are thought to be a back channel of communication regulating expression levels between paralogous genes. Given the name "competing endogenous RNAs" (ceRNAs), these microRNAs bind to "microRNA response elements" on genes and pseudogenes and may provide another explanation for the persistence of non-coding DNA.[84]
Evolution
MicroRNAs are significant phylogenetic markers because of their astonishingly low rate of evolution.[85] MicroRNAs origin as a regulatory mechanism developed from previous RNAi machinery which was initially used as a defense against exogenous genetic material such as viruses. [86] Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs[87] and perhaps, ultimately, complex life.[88] Indeed, rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.[85][87]
New microRNAs are created in multiple different ways. Novel microRNAs can originate from the random formation of hairpins in "non-coding" sections of DNA (i.e. introns or intergene regions), but also by the duplication and modification of existing microRNAs.[89]. MicroRNAs can also form from inverted duplications of protein-coding sequences, which allows for the creation of a foldback hairpin structure. [90] The rate of evolution (i.e. nucleotide substitution) in recently originated microRNAs is comparable to that elsewhere in the non-coding DNA, implying evolution by neutral drift; however, older microRNAs have a much lower rate of change (often less than one substitution per hundred million years),[88] suggesting that once a microRNA gains a function it undergoes extreme purifying selection.[89] Additionally, different regions within an miRNA gene seem to be under different evolutionary pressures, where regions that are vital for processing and function have much higher levels of conservation. [91] At this point, a microRNA is rarely lost from an animal's genome,[88] although microRNAs which are more recently derived (and thus presumably non-functional) are frequently lost.[89] In Arabidopsis thaliana, the net flux of miRNA genes has been predicted to be between 1.2 and 3.3 genes per million years. [92] This makes them a valuable phylogenetic marker, and they are being looked upon as a possible solution to such outstanding phylogenetic problems as the relationships of arthropods.[93]
MicroRNAs feature in the genomes of most eukaryotic organisms, from the brown algae[94] to the animals. However, the difference in how these microRNAs function and the way they are processed suggests that microRNAs arose independently in plants and animals. [95] Across all species, in excess of 5000 had been identified by March 2010.[96] Whilst short RNA sequences (50 – hundreds of base pairs) of a broadly comparable function occur in bacteria, bacteria lack true microRNAs.[97]
Experimental detection and manipulation of miRNA
While researchers have focused on the study of miRNA expression in physiological and pathological processes, various technical variables related to microRNA isolation have emerged. The stability of the stored miRNA samples has often been questioned.[98] MicroRNAs are degraded much more easily than mRNAs, partly due to their length, but also because of the ubiquitously present RNases. This makes it necessary to cool samples on ice and use RNase-free equipment whenever working with microRNAs.[99]
MicroRNA expression can be quantified in a two-step polymerase chain reaction process of modified RT-PCR followed by quantitative PCR. Variations of this method achieve absolute or relative quantification.[100] miRNAs can also be hybridized to microarrays, slides or chips with probes to hundreds or thousands of miRNA targets, so that relative levels of miRNAs can be determined in different samples.[101] MicroRNAs can be both discovered and profiled by high-throughput sequencing methods (MicroRNA Sequencing).[102] The activity of an miRNA can be experimentally inhibited using a locked nucleic acid (LNA) oligo, a Morpholino oligo[103][104] or a 2'-O-methyl RNA oligo.[105] Additionally, a specific miRNA can be silenced by a complementary antagomir. MicroRNA maturation can be inhibited at several points by steric-blocking oligos.[106] The miRNA target site of an mRNA transcript can also be blocked by a steric-blocking oligo.[107] For the “in situ” detection of miRNA, LNA[108] or Morpholino[109] probes can be used. The locked conformation of LNA results in enhanced hybridization properties and increases sensitivity and selectivity, making it ideal for detection of short miRNA.[110]
High-throughput quantification of miRNAs is often difficult and prone to errors, for the larger variance (compared to mRNAs) that comes with the methodological problems. mRNA-expression is therefore often analyzed as well to check for miRNA-effects in their levels (e. g. in [111][112]). To pair mRNA- and miRNA-data, databases can be used which predict miRNA-targets based on their base sequence.[113][114] While this is usually done after miRNAs of interest have been detected (e. g. because of high expression levels), ideas for analysis tools that integrate mRNA- and miRNA-expression information have been proposed.[115][116]
miRNA and disease
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease.[117] A manually curated, publicly available database, miR2Disease, documents known relationships between miRNA dysregulation and human disease.[118]
miRNA and inherited diseases
A mutation in the seed region of miR-96 causes hereditary progressive hearing loss.[119] A mutation in the seed region of miR-184 causes hereditary keratoconus with anterior polar cataract.[120] Deletion of the miR-17~92 cluster causes skeletal and growth defects.[121]
miRNA and cancer
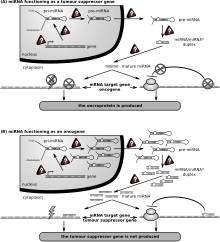
The first human disease known to be associated with miRNA deregulation was chronic lymphocytic leukemia [117] and later many miRNAs have been found to have links with some types of cancer[117][122][123] and are sometimes referred to as "oncomirs". MicroRNA-21 is involved in several cancer-types such as glioblastoma and astrocytoma was one of the first microRNAs to be identified as an oncomir.[124]
A study of mice altered to produce excess c-Myc — a protein with mutated forms implicated in several cancers — shows that miRNA has an effect on the development of cancer. Mice that were engineered to produce a surplus of types of miRNA found in lymphoma cells developed the disease within 50 days and died two weeks later. In contrast, mice without the surplus miRNA lived over 100 days.[122] Leukemia can be caused by the insertion of a viral genome next to the 17-92 array of microRNAs leading to increased expression of this microRNA.[125]
Another study found that two types of miRNA inhibit the E2F1 protein, which regulates cell proliferation. miRNA appears to bind to messenger RNA before it can be translated to proteins that switch genes on and off.[126]
By measuring activity among 217 genes encoding miRNA, patterns of gene activity that can distinguish types of cancers can be discerned. miRNA signatures may enable classification of cancer. This will allow doctors to determine the original tissue type which spawned a cancer and to be able to target a treatment course based on the original tissue type.[127] miRNA profiling has already been able to determine whether patients with chronic lymphocytic leukemia had slow growing or aggressive forms of the cancer.[123]
Transgenic mice that over-express or lack specific miRNAs have provided insight into the role of small RNAs in various malignancies.[128] Much work has also been done on the role of microRNAs in establishing and maintaining cancer stem cells that are especially resistant to chemotherapy and often responsible for relapse.[129]
A novel miRNA-profiling based screening assay for the detection of early-stage colorectal cancer has been developed and is currently in clinical trials. Early results showed that blood plasma samples collected from patients with early, resectable (Stage II) colorectal cancer could be distinguished from those of sex-and age-matched healthy volunteers. Sufficient selectivity and specificity could be achieved using small (less than 1 mL) samples of blood. The test has potential to be a cost-effective, non-invasive way to identify at-risk patients who should undergo colonoscopy.[130][131]
Another role for miRNA in cancers is to use their expression level as a prognostic, for example one study on NSCLC samples found that low miR-324a levels could serve as a prognostic indicator of poor survival,[132] another found that either high miR-185 or low miR-133b levels correlated with metastasis and poor survival in colorectal cancer.[133]
Recent studies have miR-205 targeted for inhibiting the metastatic nature of breast cancer.[134] Five members of the microRNA-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) are down regulated in tumour progression of breast cancer.[135]
miRNA, DNA repair and cancer
Individuals with an inherited deficiency in DNA repair capability are at increased risk of cancer[136] (also see DNA repair-deficiency disorder). If repair is deficient, damage tends to accumulate in DNA. Such DNA damage can cause mutational errors during DNA replication due to error-prone translesion synthesis. Accumulated DNA damage can also cause epigenetic alterations due to errors during DNA repair.[137][138] Such mutations and epigenetic alterations can give rise to cancer (see malignant neoplasms).
Germ line mutations in DNA repair genes cause only 2–5% of colon cancer cases.[139] However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important causal factor for these cancers.
Among 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1, most were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene.[140] However, up to 15% of the MLH1-deficiencies in sporadic colon cancers appeared to be due to over-expression of the microRNA miR-155, which represses MLH1 expression.[141]
In 29–66%[142][143] of glioblastomas, DNA repair is deficient due to epigenetic methylation of the MGMT gene, which reduces protein expression of MGMT. However, for 28% of glioblastomas, the MGMT protein is deficient but the MGMT promoter is not methylated.[142] In the glioblastomas without methylated MGMT promoters, the level of microRNA miR-181d is inversely correlated with protein expression of MGMT and the direct target of miR-181d is the MGMT mRNA 3’UTR (the three prime untranslated region of MGMT mRNA).[142] Thus, in 28% of glioblastomas, increased expression of miR-181d and reduced expression of DNA repair enzyme MGMT may be a causal factor.
HMGA proteins (HMGA1a, HMGA1b and HMGA2) are implicated in cancer, and expression of these proteins is regulated by microRNAs. HMGA expression is almost undetectable in differentiated adult tissues but is elevated in many cancers. HGMA proteins are polypeptides of ~100 amino acid residues characterized by a modular sequence organization. These proteins have three highly positively-charged regions, termed AT hooks, that bind the minor groove of AT-rich DNA stretches in specific regions of DNA. Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinoma, show a strong increase of HMGA1a and HMGA1b proteins.[144] Transgenic mice with HMGA1 targeted to lymphoid cells develop aggressive lymphoma, showing that high HMGA1 expression is not only associated with cancers, but that the HMGA1 gene can act as an oncogene to cause cancer.[145] Baldassarre et al.,[146] showed that HMGA1 protein binds to the promoter region of DNA repair gene BRCA1 and inhibits BRCA1 promoter activity. They also showed that while only 11% of breast tumors had hypermethylation of the BRCA1 gene, 82% of aggressive breast cancers have low BRCA1 protein expression, and most of these reductions were due to chromatin remodeling by high levels of HMGA1 protein.
HMGA2 protein specifically targets the promoter of ERCC1, thus reducing expression of this DNA repair gene.[147] ERCC1 protein expression was deficient in 100% of 47 evaluated colon cancers (though the extent to which HGMA2 was involved is not known).[148] Palmieri et al.[149] showed that, in normal tissues, HGMA1 and HMGA2 genes are targeted (and thus strongly reduced in expression) by miR-15, miR-16, miR-26a, miR-196a2 and Let-7a. However, each of these HMGA-targeting miRNAs are drastically reduced in almost all human pituitary adenomas studied, when compared with the normal pituitary gland. Consistent with the down-regulation of these HMGA-targeting miRNAs, an increase in the HMGA1 and HMGA2-specific mRNAs was observed. Three of these microRNAs (miR-16, miR-196a and Let-7a)[150][151] have methylated promoters and therefore low expression in colon cancer. For two of these, miR-15 and miR-16, the coding regions are epigenetically silenced in cancer due to histone deacetylase activity.[152] When these microRNAs are expressed at a low level, then HMGA1 and HMGA2 proteins are expressed at a high level. HMGA1 and HMGA2 target (reduce expression of) BRCA1 and ERCC1 DNA repair genes. Thus DNA repair can be reduced, likely contributing to cancer progression.[153]
In contrast to the previous example, where under-expression of miRNAs indirectly caused reduced expression of DNA repair genes, in some cases over-expression of certain miRNAs may directly reduce expression of specific DNA repair proteins. Wan et al.[154] referred to 6 DNA repair genes that are directly targeted by the miRNAs indicated: ATM (miR-421), RAD52 (miR-210, miR-373), RAD23B (miR-373), MSH2 (miR-21), BRCA1 (miR-182) and P53 (miR-504, miR-125b). Three of these miRNAs (miR-21, miR-182, miR-125b) are among those identified by Schnekenburger and Diederich[151] as over-expressed in colon cancer through epigenetic hypomethylation. Over expression of any one of these miRNAs can cause reduced expression of its target DNA repair gene.
miRNA and heart disease
The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart, and has revealed that miRNAs play an essential role during its development.[155][156] miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathies.[157][158][159] Furthermore, studies on specific miRNAs in animal models have identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response, and cardiac conductance.[156][160][161][162][163][164]
miRNA and the nervous system
miRNAs appear to regulate the nervous system.[165] Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124), synapse formation and synapse maturation (where miR-134 and miR-138 are thought to be involved).[166] Some studies find altered miRNA expression in schizophrenia.[167][168]
miRNA and obesity
miRNAs play crucial roles in the regulation of stem cell progenitors differentiating into adipocytes.[169] Studies to determine what role pluripotent stem cells play in adipogenesis, were examined in the immortalized human bone marrow-derived stromal cell line hMSC-Tert20.[170] Decreased expression of miR-155,miR-221,and miR-222, have been found during the adipogenic programming of both immortalized and primary hMSCs, suggesting that they act as negative regulators of differentiation. Conversely, ectopic expression of the miRNAs 155,221, and 222 significantly inhibited adipogenesis and repressed induction of the master regulators PPARγ and CCAAT/enhancer-binding protein alpha (CEBPA).[171] This paves the way for possible obesity treatments on the genetic level.
Another class of miRNAs that regulate insulin resistance, obesity, and diabetes, is the let-7 family. Let-7 is known to accumulate in human tissues during the course of aging. When let-7 was ectopically overexpressed to mimic accelerated aging, mice became insulin-resistant, and thus more prone to high fat diet-induced obesity and diabetes.[172] In contrast when let-7 was inhibited by injections of let-7-specific antagomirs, mice become more insulin-sensitive, and remarkably resistant to high fat diet-induced obesity and diabetes. Not only could let-7 inhibition prevent obesity and diabetes, it could also reverse and cure diabetes.[173] These experimental findings suggest that let-7 inhibition could represent a new therapy for obesity and type 2 diabetes.
miRNA and non-coding RNAs
When the human genome project mapped its first chromosome in 1999, it was predicted the genome would contain over 100,000 protein coding genes. However, only around 20,000 were eventually identified (International Human Genome Sequencing Consortium, 2004).[174] Since then, the advent of bioinformatics approaches combined with genome tiling studies examining the transcriptome,[175] systematic sequencing of full length cDNA libraries,[176] and experimental validation[177] (including the creation of miRNA derived antisense oligonucleotides called antagomirs) have revealed that many transcripts are non protein-coding RNA, including several snoRNAs and miRNAs.[178]
miRNA and viruses
The expression of transcription activators by human herpesvirus-6 DNA is believed to be regulated by viral miRNA.[179]
See also
- Gene expression
- List of miRNA gene prediction tools
- List of miRNA target prediction tools
- RNAi
- siRNA
- Small nucleolar RNA derived microRNA
References
- ^ Chen, Kevin; Rajewsky, Nikolaus (2007). "The evolution of gene regulation by transcription factors and microRNAs". Nature Reviews Genetics. 8 (2): 93–103. doi:10.1038/nrg1990. PMID 17230196.
- ^ Bartel DP (2009). "MicroRNAs: target recognition and regulatory functions". Cell. 136 (2): 215–33. doi:10.1016/j.cell.2009.01.002. PMID 19167326.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Kusenda B, Mraz M, Mayer J, Pospisilova S (2006). "MicroRNA biogenesis, functionality and cancer relevance". Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 150 (2): 205–15. doi:10.5507/bp.2006.029. PMID 17426780.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Homo sapiens miRNAs in the miRBase at Manchester University
- ^ Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z (2005). "Identification of hundreds of conserved and nonconserved human microRNAs". Nat. Genet. 37 (7): 766–70. doi:10.1038/ng1590. PMID 15965474.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Lewis BP, Burge CB, Bartel DP (2005). "Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets". Cell. 120 (1): 15–20. doi:10.1016/j.cell.2004.12.035. PMID 15652477.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Friedman RC, Farh KK, Burge CB, Bartel DP (2009). "Most mammalian mRNAs are conserved targets of microRNAs". Genome Res. 19 (1): 92–105. doi:10.1101/gr.082701.108. PMC 2612969. PMID 18955434.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP (2003). "The microRNAs of Caenorhabditis elegans". Genes Dev. 17 (8): 991–1008. doi:10.1101/gad.1074403. PMC 196042. PMID 12672692.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tanzer A, Stadler PF (2004). "Molecular evolution of a microRNA cluster". J. Mol. Biol. 339 (2): 327–35. doi:10.1016/j.jmb.2004.03.065. PMID 15136036.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC (2007). "miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii". Nature. 447 (7148): 1126–9. Bibcode:2007Natur.447.1126M. doi:10.1038/nature05903. PMID 17538623.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ (2009). "MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis". RNA Biol. 6 (1): 65–72. doi:10.4161/rna.6.1.7534. PMID 19106625.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lee CT, Risom T, Strauss WM (2007). "Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny". DNA Cell Biol. 26 (4): 209–18. doi:10.1089/dna.2006.0545. PMID 17465887.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Shabalina SA, Koonin EV (2008). "Origins and evolution of eukaryotic RNA interference". Trends in Ecology and Evolution. 10 (10): 578–587. doi:10.1016/j.tree.2008.06.005. PMC 2695246. PMID 18715673.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008). "Widespread translational inhibition by plant miRNAs and siRNAs". Science. 320 (5880): 1185–90. Bibcode:2008Sci...320.1185B. doi:10.1126/science.1159151. PMID 18483398.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b He L, Hannon GJ (2004). "MicroRNAs: small RNAs with a big role in gene regulation". Nature. 5 (7): 522–531. doi:10.1038/nrg1379. PMID 15211354.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Lewis BP, Shih IH, Jones-Rhoades M, Bartel DP, Burge CB (2003). "Prediction of Mammalian MicroRNA Targets". Cell. 115 (7): 787–798. doi:10.1016/S0092-8674(03)01018-3. PMID 14697198.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rajewsky, Nikolaus (NaN undefined NaN). "microRNA target predictions in animals". Nature Genetics. 38 (6s): S8–S13. doi:10.1038/ng1798.
{{cite journal}}: Check date values in:|date=(help) - ^ Krek, Azra (NaN undefined NaN). "Combinatorial microRNA target predictions". Nature Genetics. 37 (5): 495–500. doi:10.1038/ng1536. PMID 15806104.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Lee RC, Feinbaum RL, Ambros V (1993). "The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14". Cell. 75 (5): 843–54. doi:10.1016/0092-8674(93)90529-Y. PMID 8252621.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005). "Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs". Nature. 433 (7027): 769–73. Bibcode:2005Natur.433..769L. doi:10.1038/nature03315. PMID 15685193.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) Cite error: The named reference "pmid15685193" was defined multiple times with different content (see the help page). - ^ Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003). "bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila". Cell. 113 (1): 25–36. doi:10.1016/S0092-8674(03)00231-9. PMID 12679032.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Cuellar TL, McManus MT (2005). "MicroRNAs and endocrine biology". J. Endocrinol. 187 (3): 327–32. doi:10.1677/joe.1.06426. PMID 16423811.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M (2004). "A pancreatic islet-specific microRNA regulates insulin secretion". Nature. 432 (7014): 226–30. Bibcode:2004Natur.432..226P. doi:10.1038/nature03076. PMID 15538371.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Chen CZ, Li L, Lodish HF, Bartel DP (2004). "MicroRNAs modulate hematopoietic lineage differentiation". Science. 303 (5654): 83–6. Bibcode:2004Sci...303...83C. doi:10.1126/science.1091903. PMID 14657504.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Wilfred BR, Wang WX, Nelson PT (2007). "Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways". Mol. Genet. Metab. 91 (3): 209–17. doi:10.1016/j.ymgme.2007.03.011. PMC 1978064. PMID 17521938.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ (2005). "The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb". Proc. Natl. Acad. Sci. U.S.A. 102 (31): 10898–903. Bibcode:2005PNAS..10210898H. doi:10.1073/pnas.0504834102. PMC 1182454. PMID 16040801.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002). "Identification of tissue-specific microRNAs from mouse". Curr. Biol. 12 (9): 735–9. doi:10.1016/S0960-9822(02)00809-6. PMID 12007417.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Trang P, Weidhaas JB, Slack FJ (2008). "MicroRNAs as potential cancer therapeutics". Oncogene. 27 Suppl 2: S52–7. doi:10.1038/onc.2009.353. PMID 19956180.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Li C, Feng Y, Coukos G, Zhang L (2009). "Therapeutic microRNA strategies in human cancer". AAPS J. 11 (4): 747–57. doi:10.1208/s12248-009-9145-9. PMC 2782079. PMID 19876744.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F (2010). "microRNA: emerging therapeutic targets in acute ischemic diseases". Pharmacol. Ther. 125 (1): 92–104. doi:10.1016/j.pharmthera.2009.10.003. PMID 19896977.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hydbring, Per (2013). "Clinical applications of microRNAs". F1000Research. 2. doi:10.12688/f1000research.2-136.v2.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000). "The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans". Nature. 403 (6772): 901–6. Bibcode:2000Natur.403..901R. doi:10.1038/35002607. PMID 10706289.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G (2000). "Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA". Nature. 408 (6808): 86–9. doi:10.1038/35040556. PMID 11081512.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003). "A uniform system for microRNA annotation". RNA. 9 (3): 277–9. doi:10.1261/rna.2183803. PMC 1370393. PMID 12592000.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006). "miRBase: microRNA sequences, targets and gene nomenclature". Nucleic Acids Res. 34 (Database issue): D140–4. doi:10.1093/nar/gkj112. PMC 1347474. PMID 16381832.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Lau NC, Lim LP, Weinstein EG, Bartel DP (2001). "An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans". Science. 294 (5543): 858–62. Bibcode:2001Sci...294..858L. doi:10.1126/science.1065062. PMID 11679671.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d e f Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004). "MicroRNA genes are transcribed by RNA polymerase II". EMBO J. 23 (20): 4051–60. doi:10.1038/sj.emboj.7600385. PMC 524334. PMID 15372072.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001). "Identification of novel genes coding for small expressed RNAs". Science. 294 (5543): 853–8. Bibcode:2001Sci...294..853L. doi:10.1126/science.1064921. PMID 11679670.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lee RC, Ambros V (2001). "An extensive class of small RNAs in Caenorhabditis elegans". Science. 294 (5543): 862–4. Bibcode:2001Sci...294..862L. doi:10.1126/science.1065329. PMID 11679672.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B, Pavlova S, Borsky M, Verner J, Doubek M, Brychtova Y, Trbusek M, Hampl A, Mayer J, Pospisilova S (2012). "MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia". Blood. 119 (9): 2110–3. doi:10.1182/blood-2011-11-394874. PMID 22234685.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004). "Identification of mammalian microRNA host genes and transcription units". Genome Res. 14 (10A): 1902–10. doi:10.1101/gr.2722704. PMC 524413. PMID 15364901.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d Cai X, Hagedorn CH, Cullen BR (2004). "Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs". RNA. 10 (12): 1957–66. doi:10.1261/rna.7135204. PMC 1370684. PMID 15525708.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Weber MJ (2005). "New human and mouse microRNA genes found by homology search". FEBS J. 272 (1): 59–73. doi:10.1111/j.1432-1033.2004.04389.x. PMID 15634332.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Kim YK, Kim VN (2007). "Processing of intronic microRNAs". EMBO J. 26 (3): 775–83. doi:10.1038/sj.emboj.7601512. PMC 1794378. PMID 17255951.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Baskerville S, Bartel DP (2005). "Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes". RNA. 11 (3): 241–7. doi:10.1261/rna.7240905. PMC 1370713. PMID 15701730.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H (2005). "Clustering and conservation patterns of human microRNAs". Nucleic Acids Res. 33 (8): 2697–706. doi:10.1093/nar/gki567. PMC 1110742. PMID 15891114.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Zhou X, Ruan J, Wang G, Zhang W (2007). "Characterization and identification of microRNA core promoters in four model species". PLoS Comput. Biol. 3 (3): e37. Bibcode:2007PLSCB...3...37Z. doi:10.1371/journal.pcbi.0030037. PMC 1817659. PMID 17352530.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Faller M, Guo F (2008). "MicroRNA biogenesis: there's more than one way to skin a cat". Biochim. Biophys. Acta. 1779 (11): 663–7. doi:10.1016/j.bbagrm.2008.08.005. PMC 2633599. PMID 18778799.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Gregory RI, Chendrimada TP, Shiekhattar R (2006). "MicroRNA biogenesis: isolation and characterization of the microprocessor complex". Methods Mol. Biol. 342: 33–47. doi:10.1385/1-59745-123-1:33. ISBN 1-59745-123-1. PMID 16957365.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC (2007). "Mammalian mirtron genes". Mol. Cell. 28 (2): 328–36. doi:10.1016/j.molcel.2007.09.028. PMC 2763384. PMID 17964270.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K (2008). "Frequency and fate of microRNA editing in human brain". Nucleic Acids Res. 36 (16): 5270–80. doi:10.1093/nar/gkn479. PMC 2532740. PMID 18684997.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009). "Many roads to maturity: microRNA biogenesis pathways and their regulation". Nat. Cell Biol. 11 (3): 228–34. doi:10.1038/ncb0309-228. PMID 19255566.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ohman M (2007). "A-to-I editing challenger or ally to the microRNA process". Biochimie. 89 (10): 1171–6. doi:10.1016/j.biochi.2007.06.002. PMID 17628290.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Murchison EP, Hannon GJ (2004). "miRNAs on the move: miRNA biogenesis and the RNAi machinery". Curr. Opin. Cell Biol. 16 (3): 223–9. doi:10.1016/j.ceb.2004.04.003. PMID 15145345.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Lund E, Dahlberg JE (2006). "Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs". Cold Spring Harb. Symp. Quant. Biol. 71: 59–66. doi:10.1101/sqb.2006.71.050. PMID 17381281.
- ^ Ji X (2008). "The mechanism of RNase III action: how dicer dices". Curr. Top. Microbiol. Immunol. Current Topics in Microbiology and Immunology. 320: 99–116. doi:10.1007/978-3-540-75157-1_5. ISBN 978-3-540-75156-4. PMID 18268841.
- ^ Lelandais-Brière C, Sorin C, Declerck M, Benslimane A, Crespi M, Hartmann C (2010). "Small RNA diversity in plants and its impact in development". Current Genomics. 11 (1): 14–23. doi:10.2174/138920210790217918. PMC 2851111. PMID 20808519.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rana TM (2007). "Illuminating the silence: understanding the structure and function of small RNAs". Nat. Rev. Mol. Cell Biol. 8 (1): 23–36. doi:10.1038/nrm2085. PMID 17183358.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Schwarz DS, Zamore PD (2002). "Why do miRNAs live in the miRNP?". Genes Dev. 16 (9): 1025–31. doi:10.1101/gad.992502. PMID 12000786.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ (2004). "Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design". J Biol Chem. 279 (40): 42230–9. doi:10.1074/jbc.M404931200. PMID 15292246.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Khvorova A, Reynolds A, Jayasena SD (2003). "Functional siRNAs and miRNAs exhibit strand bias". Cell. 115 (2): 209–16. doi:10.1016/S0092-8674(03)00801-8. PMID 14567918.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD (2003). "Asymmetry in the assembly of the RNAi enzyme complex". Cell. 115 (2): 199–208. doi:10.1016/S0092-8674(03)00759-1. PMID 14567917.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lin SL, Chang D, Ying SY (2005). "Asymmetry of intronic pre-miRNA structures in functional RISC assembly". Gene. 356: 32–8. doi:10.1016/j.gene.2005.04.036. PMC 1788082. PMID 16005165.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Okamura K, Chung WJ, Lai EC (2008). "The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs". Cell Cycle. 7 (18): 2840–5. doi:10.4161/cc.7.18.6734. PMC 2697033. PMID 18769156.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Pratt AJ, MacRae IJ (2009). "The RNA-induced silencing complex: a versatile gene-silencing machine". J. Biol. Chem. 284 (27): 17897–901. doi:10.1074/jbc.R900012200. PMC 2709356. PMID 19342379.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA (2008). "In vitro reconstitution of the human RISC-loading complex". Proc. Natl. Acad. Sci. U.S.A. 105 (2): 512–7. Bibcode:2008PNAS..105..512M. doi:10.1073/pnas.0710869105. PMC 2206567. PMID 18178619.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002). "miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs". Genes Dev. 16 (6): 720–8. doi:10.1101/gad.974702. PMC 155365. PMID 11914277.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Meister G, Landthaler M, Peters L, Chen P, Urlaub H, Lurhmann R, Tuschl T (2005). "Identification of Novel Argonaute-Associated Proteins". Current Biology. 15 (23): 2149–55. doi:10.1016/j.cub.2005.10.048. PMID 16289642.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Kai ZS, Pasquinelli AE (2010). "MicroRNA assassins: factors that regulate the disappearance of miRNAs". Nat. Struct. Mol. Biol. 17 (1): 5–10. doi:10.1038/nsmb.1762. PMID 20051982.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Chatterjee S, Großhans H (2009). "Active turnover modulates mature microRNA activity in Caenorhabditis elegans". Nature. 461 (7263): 546–459. Bibcode:2009Natur.461..546C. doi:10.1038/nature08349. PMID 19734881.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Morozova N, Zinovyev A, Nonne N, Pritchard LL, Gorban AN, Harel-Bellan A (2012). "Kinetic signatures of microRNA modes of action". RNA. 18 (9): 1635–55. doi:10.1261/rna.032284.112. PMC 3425779. PMID 22850425.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004). "Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets". Genome Biol. 5 (9): R65. doi:10.1186/gb-2004-5-9-r65. PMC 522872. PMID 15345049.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kawasaki H, Taira K (2004). "MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells". Nucleic Acids Symp Ser. 48 (48): 211–2. doi:10.1093/nass/48.1.211. PMID 17150553.
- ^ a b Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008). "Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening". Genome Res. 18 (10): 1602–9. doi:10.1101/gr.080127.108. PMC 2556272. PMID 18653800.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Mazière P, Enright AJ (2007). "Prediction of microRNA targets". Drug Discov. Today. 12 (11–12): 452–8. doi:10.1016/j.drudis.2007.04.002. PMID 17532529.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Williams AE (2008). "Functional aspects of animal microRNAs". Cell. Mol. Life Sci. 65 (4): 545–62. doi:10.1007/s00018-007-7355-9. PMID 17965831.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009). "Deadenylation is a widespread effect of miRNA regulation". RNA. 15 (1): 21–32. doi:10.1261/rna.1399509. PMC 2612776. PMID 19029310.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bazzini AA, Lee MT, Giraldez AJ (2012). "Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish". Science. 336 (6078): 233–7. Bibcode:2012Sci...336..233B. doi:10.1126/science.1215704. PMID 22422859.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R (2009). "Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells". BMC Mol. Biol. 10: 12. doi:10.1186/1471-2199-10-12. PMC 2680403. PMID 19232136.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Hawkins PG, Morris KV (2008). "RNA and transcriptional modulation of gene expression". Cell Cycle. 7 (5): 602–7. doi:10.4161/cc.7.5.5522. PMC 2877389. PMID 18256543.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005). "Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution". Cell. 123 (6): 1133–46. doi:10.1016/j.cell.2005.11.023. PMID 16337999.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Li LC (2008). "Small RNA-Mediated Gene Activation". RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN [[Special:BookSources/978-1-904455-25-7]|978-1-904455-25-7]]]. http://www.horizonpress.com/rnareg.
{{cite book}}: Check|isbn=value: invalid character (help); External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help)CS1 maint: extra punctuation (link) - ^ Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008). "MicroRNA-373 induces expression of genes with complementary promoter sequences". Proc. Natl. Acad. Sci. U.S.A. 105 (5): 1608–13. Bibcode:2008PNAS..105.1608P. doi:10.1073/pnas.0707594105. PMC 2234192. PMID 18227514.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011). "A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?". Cell. 146 (3): 353–8. doi:10.1016/j.cell.2011.07.014. PMC 3235919. PMID 21802130.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ (2009). "The deep evolution of metazoan microRNAs". Evol. Dev. 11 (1): 50–68. doi:10.1111/j.1525-142X.2008.00302.x. PMID 19196333.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pashkovskiy, P. P. (2013). "Biogenesis, evolution, and functions of plant microRNAs". Biochemistry-Moscow. 78: 627–637. doi:10.1134/S0006297913060084. PMID 23980889.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ (2008). "MicroRNAs and the advent of vertebrate morphological complexity". Proc. Natl. Acad. Sci. U.S.A. 105 (8): 2946–50. Bibcode:2008PNAS..105.2946H. doi:10.1073/pnas.0712259105. PMC 2268565. PMID 18287013.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Peterson KJ, Dietrich MR, McPeek MA (2009). "MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion". BioEssays. 31 (7): 736–47. doi:10.1002/bies.200900033. PMID 19472371.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Nozawa M, Miura S, Nei M (2010). "Origins and evolution of microRNA genes in Drosophila species". Genome Biol Evol. 2: 180–9. doi:10.1093/gbe/evq009. PMC 2942034. PMID 20624724.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Allen, E. (2004). "Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana". Nature Genetics. 36: 1282–1290. doi:10.1038/ng1478.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Warthmann, N. (2008). "Comparative analysis of the MIR319a MicroRNA locus in Arabidopsis and related Brassicaceae". Molecular Biology and Evolution. 25 (5): 892–902. doi:10.1093/molbev/msn029. PMID 18296705.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Fahlgren, N. (2010). "MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana". Plant Cell. 22 (4): 1074–1089. doi:10.1105/tpc.110.073999.
{{cite journal}}: Check|doi=value (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); zero width space character in|doi=at position 4 (help) - ^ Caravas J, Friedrich M (2010). "Of mites and millipedes: recent progress in resolving the base of the arthropod tree". BioEssays. 32 (6): 488–95. doi:10.1002/bies.201000005. PMID 20486135.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH; et al. (2010). "The Ectocarpus genome and the independent evolution of multicellularity in brown algae". Nature. 465 (7298): 617–21. Bibcode:2010Natur.465..617C. doi:10.1038/nature09016. PMID 20520714.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Cuperus, J. T. (2011). "Evolution and functional diversification of MIRNA genes". Plant Cell. 23 (2): 431–442. doi:10.1105/tpc.110.082784. PMID 21317375.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Dimond PF (15 March 2010). "miRNAs' Therapeutic Potential". Genetic Engineering & Biotechnology News. Vol. 30, no. 6. p. 1. Archived from the original on 10 July 2010. Retrieved 10 July 2010.
- ^ Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G (2006). "Target prediction for small, noncoding RNAs in bacteria". Nucleic Acids Res. 34 (9): 2791–802. doi:10.1093/nar/gkl356. PMC 1464411. PMID 16717284.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mraz M, Malinova K, Mayer J, Pospisilova S (2009). "MicroRNA isolation and stability in stored RNA samples". Biochem. Biophys. Res. Commun. 390 (1): 1–4. doi:10.1016/j.bbrc.2009.09.061. PMID 19769940.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Liu CG, Calin GA, Volinia S, Croce CM (2008). "MicroRNA expression profiling using microarrays". Nat Protoc. 3 (4): 563–78. doi:10.1038/nprot.2008.14. PMID 18388938.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005). "Real-time quantification of microRNAs by stem-loop RT-PCR". Nucleic Acids Res. 33 (20): e179. doi:10.1093/nar/gni178. PMC 1292995. PMID 16314309.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E (2005). "An optimized isolation and labeling platform for accurate microRNA expression profiling". RNA. 11 (9): 1461–70. doi:10.1261/rna.2610405. PMC 1370829. PMID 16043497.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, 't Hoen PA. (2010). "New methods for next generation sequencing based microRNA expression profiling". BMC Genomics. 11: 716. doi:10.1186/1471-2164-11-716. PMC 3022920. PMID 21171994.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH (2004). "Substrate requirements for let-7 function in the developing zebrafish embryo". Nucleic Acids Res. 32 (21): 6284–91. doi:10.1093/nar/gkh968. PMC 535676. PMID 15585662.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (2007). "Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate". Nat. Genet. 39 (2): 259–63. doi:10.1038/ng1953. PMID 17220889.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Meister G, Landthaler M, Dorsett Y, Tuschl T (2004). "Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing". RNA. 10 (3): 544–50. doi:10.1261/rna.5235104. PMC 1370948. PMID 14970398.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH (2007). "Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development". PLoS Biol. 5 (8): e203. doi:10.1371/journal.pbio.0050203. PMC 1925136. PMID 17676975.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Choi WY, Giraldez AJ, Schier AF (2007). "Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430". Science. 318 (5848): 271–4. Bibcode:2007Sci...318..271C. doi:10.1126/science.1147535. PMID 17761850.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ You Y, Moreira BG, Behlke MA, Owczarzy R (2006). "Design of LNA probes that improve mismatch discrimination". Nucleic Acids Res. 34 (8): e60. doi:10.1093/nar/gkl175. PMC 1456327. PMID 16670427.
{{cite journal}}: Cite has empty unknown parameter:|month=(help)CS1 maint: multiple names: authors list (link) - ^ Lagendijk AK, Moulton JD, Bakkers J (2012). "Revealing details: whole mount microRNA in situ hybridization protocol for zebrafish embryos and adult tissues". Bio Open. 1 (6): 566. doi:10.1242/bio.2012810.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kaur H, Arora A, Wengel J, Maiti S, Arora A, Wengel J, Maiti S (2006). "Thermodynamic, Counterion, and Hydration Effects for the Incorporation of Locked Nucleic Acid Nucleotides into DNA Duplexes". Biochemistry. 45 (23): 7347–55. doi:10.1021/bi060307w. PMID 16752924.
{{cite journal}}: Cite has empty unknown parameter:|month=(help)CS1 maint: multiple names: authors list (link) - ^ Nielsen JA, Lau P, Maric D, Barker JL, Hudson LD (2009). "Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis". BMC Neurosci. 10: 98. doi:10.1186/1471-2202-10-98. PMC 2736963. PMID 19689821.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, Bar-Joseph Z, Mathavan S, Ayyavoo V (2011). Mammano, Fabrizio (ed.). "Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1)". PLoS ONE. 6 (7): e22730. doi:10.1371/journal.pone.0022730. PMC 3145673. PMID 21829495.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007). "MicroRNA targeting specificity in mammals: determinants beyond seed pairing". Mol. Cell. 27 (1): 91–105. doi:10.1016/j.molcel.2007.06.017. PMID 17612493.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008). "miRBase: tools for microRNA genomics". Nucleic Acids Res. 36 (Database issue): D154–8. doi:10.1093/nar/gkm952. PMC 2238936. PMID 17991681.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP (2009). "MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression". Nucleic Acids Res. 37 (Web Server issue): W356–62. doi:10.1093/nar/gkp294. PMC 2703907. PMID 19420067.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Artmann S, Jung K, Bleckmann A, Beissbarth T (2012). Provero, Paolo (ed.). "Detection of simultaneous group effects in microRNA expression and related target gene sets". PLoS ONE. 7 (6): e38365. Bibcode:2012PLoSO...738365A. doi:10.1371/journal.pone.0038365. PMC 3378551. PMID 22723856.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 23216588, please use {{cite journal}} with
|pmid=23216588instead. - ^ Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. (2009). "miR2Disease: a manually curated database for microRNA deregulation in human disease". Nucleic Acids Research. 37. (Database issue) (Database issue): D98–104. doi:10.1093/nar/gkn714. PMC 2686559. PMID 18927107.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA (2009). "Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss". Nat. Genet. 41 (5): 609–13. doi:10.1038/ng.355. PMID 19363479.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, Willoughby CE (2011). "Mutation Altering the miR-184 Seed Region Causes Familial Keratoconus with Cataract". The American Journal of Human Genetics. 89 (5): 628–633. doi:10.1016/j.ajhg.2011.09.014. PMC 3213395. PMID 21996275.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Geneviève D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J (2011). "Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans". Nat. Genet. 43 (10): 1026–30. doi:10.1038/ng.915. PMC 3184212. PMID 21892160.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005). "A microRNA polycistron as a potential human oncogene". Nature. 435 (7043): 828–33. Bibcode:2005Natur.435..828H. doi:10.1038/nature03552. PMID 15944707.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J (2009). "MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes". Leuk. Lymphoma. 50 (3): 506–9. doi:10.1080/10428190902763517. PMID 19347736.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3538124/
- ^ Cui JW, Li YJ, Sarkar A, Brown J, Tan YH, Premyslova M, Michaud C, Iscove N, Wang GJ, Ben-David Y. (2007). "Retroviral insertional activation of the Fli-3 locus in erythroleukemias encoding a cluster of microRNAs that convert Epo-induced differentiation to proliferation". Blood. 110 (7): 2631–40. doi:10.1182/blood-2006-10-053850. PMID 17586726.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005). "c-Myc-regulated microRNAs modulate E2F1 expression". Nature. 435 (7043): 839–43. Bibcode:2005Natur.435..839O. doi:10.1038/nature03677. PMID 15944709.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005). "MicroRNA expression profiles classify human cancers". Nature. 435 (7043): 834–8. Bibcode:2005Natur.435..834L. doi:10.1038/nature03702. PMID 15944708.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Zanesi N, Pekarsky Y, Trapasso F, Calin G, Croce CM (2010). "MicroRNAs in mouse models of lymphoid malignancies". J Nucleic Acids Investig. 1 (1): 36–40. doi:10.4081/jnai.2010.e8. PMC 3111058. PMID 21666870.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jun Qian, Vinayakumar Siragam, Jiang Lin, Jichun Ma, Zhaoqun Deng (2011). "The role of microRNAs in the formation of cancer stem cells: Future directions for miRNAs". Hypothesis. 9 (1): e10.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Screening Tool Can Detect Colorectal Cancer from a Small Blood Sample" (Press release). American Association for Cancer Research. 29 September 2010. Retrieved 29 November 2010.
- ^ Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S, Nielsen HJ (2010). "High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients". Clin Exp Metastasis. 28 (1): 27–38. doi:10.1007/s10585-010-9355-7. PMC 2998639. PMID 21069438.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Võsa U, Vooder T, Kolde R, Fischer K, Välk K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T (2011). "Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer". Genes Chromosomes Cancer. 50 (10): 812–22. doi:10.1002/gcc.20902. PMID 21748820.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Akçakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO (2011). "miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer". Int. J. Oncol. 39 (2): 311–8. doi:10.3892/ijo.2011.1043. PMID 21573504.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Wu H, Mo YY (2009). "Targeting miR-205 in breast cancer". Expert Opin. Ther. Targets. 13 (12): 1439–48. doi:10.1517/14728220903338777. PMID 19839716.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ (2008). "The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1". Nat. Cell Biol. 10 (5): 593–601. doi:10.1038/ncb1722. PMID 18376396.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bernstein C, Bernstein H, Payne CM, Garewal H. "DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002 Jun;511(2) 145-78. Review.
- ^ O'Hagan HM, Mohammad HP, Baylin SB. (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island" PLoS Genet 4(8) e1000155. doi:10.1371/journal.pgen.1000155 PMID 18704159
- ^ Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV (2007). DNA damage, homology-directed repair, and DNA methylation" PLoS Genet 3(7) e110. doi:10.1371/journal.pgen.0030110 PMID 17616978
- ^ Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010). Hereditary and familial colon cancer" Gastroenterology 138(6) 2044-2058. doi: 10.1053/j.gastro.2010.01.054. PMID 20420945
- ^ Truninger K, Menigatti M, Luz J; et al. (2005). "Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer". Gastroenterology. 128 (5): 1160–71. doi:10.1053/j.gastro.2005.01.056. PMID 15887099.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Valeri N, Gasparini P, Fabbri M; et al. (2010). "Modulation of mismatch repair and genomic stability by miR-155". Proc. Natl. Acad. Sci. U.S.A. 107 (15): 6982–7. Bibcode:2010PNAS..107.6982V. doi:10.1073/pnas.1002472107. PMC 2872463. PMID 20351277.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Zhang W, Zhang J, Hoadley K; et al. (2012). "miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression". Neuro-oncology. 14 (6): 712–9. doi:10.1093/neuonc/nos089. PMC 3367855. PMID 22570426.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Spiegl-Kreinecker S, Pirker C, Filipits M; et al. (2010). "O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients". Neuro-oncology. 12 (1): 28–36. doi:10.1093/neuonc/nop003. PMC 2940563. PMID 20150365.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Sgarra R, Rustighi A, Tessari MA; et al. (2004). "Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer". FEBS Lett. 574 (1–3): 1–8. doi:10.1016/j.febslet.2004.08.013. PMID 15358530.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Xu Y, Sumter TF, Bhattacharya R; et al. (2004). "The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia". Cancer Res. 64 (10): 3371–5. doi:10.1158/0008-5472.CAN-04-0044. PMID 15150086.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Baldassarre G, Battista S, Belletti B; et al. (2003). "Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma". Mol. Cell. Biol. 23 (7): 2225–38. doi:10.1128/MCB.23.7.2225-2238.2003. PMC 150734. PMID 12640109.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Borrmann L, Schwanbeck R, Heyduk T; et al. (2003). "High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity". Nucleic Acids Res. 31 (23): 6841–51. doi:10.1093/nar/gkg884. PMC 290254. PMID 14627817.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Facista A, Nguyen H, Lewis C, Prasad AR, Ramsey L, Zaitlin B, Nfonsam V, Krouse RS, Bernstein H, Payne CM, Stern S, Oatman N, Banerjee B, Bernstein C. (2012). Deficient expression of DNA repair enzymes in early progression to sporadic colon cancer. Genome Integr 3(1) 3. doi: 10.1186/2041-9414-3-3. PMID 22494821 http://www.ncbi.nlm.nih.gov/pubmed/22494821
- ^ Palmieri D, D'Angelo D, Valentino T; et al. (2012). "Downregulation of HMGA-targeting microRNAs has a critical role in human pituitary tumorigenesis". Oncogene. 31 (34): 3857–65. doi:10.1038/onc.2011.557. PMID 22139073.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Malumbres M (2013). "miRNAs and cancer: an epigenetics view". Mol. Aspects Med. 34 (4): 863–74. doi:10.1016/j.mam.2012.06.005. PMID 22771542.
- ^ a b Schnekenburger M, Diederich M (2012). "Epigenetics Offer New Horizons for Colorectal Cancer Prevention". Curr Colorectal Cancer Rep. 8 (1): 66–81. doi:10.1007/s11888-011-0116-z. PMC 3277709. PMID 22389639.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Sampath D, Liu C, Vasan K; et al. (2012). "Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia". Blood. 119 (5): 1162–72. doi:10.1182/blood-2011-05-351510. PMC 3277352. PMID 22096249.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bernstein C, Prasad AR, Nfonsam V, Bernstein H. (2013). DNA Damage, DNA Repair and Cancer, New Research Directions in DNA Repair, Prof. Clark Chen (Ed.), ISBN 978-953-51-1114-6, InTech, http://www.intechopen.com/books/new-research-directions-in-dna-repair/dna-damage-dna-repair-and-cancer
- ^ Wan G, Mathur R, Hu X, Zhang X, Lu X (2011). "miRNA response to DNA damage". Trends Biochem. Sci. 36 (9): 478–84. doi:10.1016/j.tibs.2011.06.002. PMC 3532742. PMID 21741842.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ (2008). "Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure". Proc. Natl. Acad. Sci. U.S.A. 105 (6): 2111–6. Bibcode:2008PNAS..105.2111C. doi:10.1073/pnas.0710228105. PMC 2542870. PMID 18256189.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D (2007). "Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2". Cell. 129 (2): 303–17. doi:10.1016/j.cell.2007.03.030. PMID 17397913.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J (2007). "MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure". Circulation. 116 (3): 258–67. doi:10.1161/CIRCULATIONAHA.107.687947. PMID 17606841.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN (2006). "A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure". Proc. Natl. Acad. Sci. U.S.A. 103 (48): 18255–60. Bibcode:2006PNAS..10318255V. doi:10.1073/pnas.0608791103. PMC 1838739. PMID 17108080.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ (2007). "Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy". J. Mol. Cell. Cardiol. 42 (6): 1137–41. doi:10.1016/j.yjmcc.2007.04.004. PMC 1934409. PMID 17498736.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Zhao Y, Samal E, Srivastava D (2005). "Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis". Nature. 436 (7048): 214–20. Bibcode:2005Natur.436..214Z. doi:10.1038/nature03817. PMID 15951802.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Zhang Y, Yang B, Wang Z (2007). "MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts". J. Biol. Chem. 282 (17): 12363–7. doi:10.1074/jbc.C700015200. PMID 17344217.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z (2007). "The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2". Nat. Med. 13 (4): 486–91. doi:10.1038/nm1569. PMID 17401374.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G (2007). "MicroRNA-133 controls cardiac hypertrophy". Nat. Med. 13 (5): 613–8. doi:10.1038/nm1582. PMID 17468766.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN (2007). "Control of stress-dependent cardiac growth and gene expression by a microRNA". Science. 316 (5824): 575–9. Bibcode:2007Sci...316..575V. doi:10.1126/science.1139089. PMID 17379774.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Maes OC, Chertkow HM, Wang E, Schipper HM (2009). "MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders". Current Genomics. 10 (3): 154–68. doi:10.2174/138920209788185252. PMC 2705849. PMID 19881909.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Schratt G (2009). "microRNAs at the synapse". Nat. Rev. Neurosci. 10 (12): 842–9. doi:10.1038/nrn2763. PMID 19888283.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Feng J, Sun G, Yan J, Noltner K, Li W, Buzin CH, Longmate J, Heston LL, Rossi J, Sommer SS (2009). Reif, Andreas (ed.). "Evidence for X-chromosomal schizophrenia associated with microRNA alterations". PLoS ONE. 4 (7): e6121. Bibcode:2009PLoSO...4.6121F. doi:10.1371/journal.pone.0006121. PMC 2699475. PMID 19568434.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ (2009). "Schizophrenia is associated with an increase in cortical microRNA biogenesis". Mol. Psychiatry. 15 (12): 1176–89. doi:10.1038/mp.2009.84. PMC 2990188. PMID 19721432.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL (2011). "MicroRNA regulation in mammalian adipogenesis". Exp. Biol. Med. (Maywood). 236 (9): 997–1004. doi:10.1258/ebm.2011.011101. PMID 21844119.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Skårn M, Namløs HM, Noordhuis P, Wang MY, Meza-Zepeda LA, Myklebost O (2012). "Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222". Stem Cells Dev. 21 (6): 873–83. doi:10.1089/scd.2010.0503. PMID 21756067.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Zuo Y, Qiang L, Farmer SR (2006). "Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter". J. Biol. Chem. 281 (12): 7960–7. doi:10.1074/jbc.M510682200. PMID 16431920.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI; DIAGRAM Consortium; MAGIC Investigators, Altshuler D, Daley GQ (2011). "The Lin28/let-7 axis regulates glucose metabolism". Cell. 147 (1): 81–94. doi:10.1016/j.cell.2011.08.033. PMC 3353524. PMID 21962509.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Frost RJ, Olson EN. (2011). "Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs". Proc Natl Acad Sci U S A. 108 (52): 21075–80. Bibcode:2011PNAS..10821075F. doi:10.1073/pnas.1118922109. PMC 3248488. PMID 22160727.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Pheasant M, Mattick JS (2007). "Raising the estimate of functional human sequences". Genome Res. 17 (9): 1245–53. doi:10.1101/gr.6406307. PMID 17690206.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M (2004). "Global identification of human transcribed sequences with genome tiling arrays". Science. 306 (5705): 2242–6. Bibcode:2004Sci...306.2242B. doi:10.1126/science.1103388. PMID 15539566.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M; et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kuhn DE, Martin MM, Feldman DS, Terry AV, Nuovo GJ, Elton TS (2008). "Experimental validation of miRNA targets". Methods. 44 (1): 47–54. doi:10.1016/j.ymeth.2007.09.005. PMC 2237914. PMID 18158132.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hüttenhofer A, Schattner P, Polacek N (2005). "Non-coding RNAs: hope or hype?". Trends Genet. 21 (5): 289–97. doi:10.1016/j.tig.2005.03.007. PMID 15851066.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S (2012). "Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B". J. Virol. 86 (3): 1638–49. doi:10.1128/JVI.05911-11. PMC 3264354. PMID 22114334.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link)
Bibliography
- miRNA definition and classification: Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003). "A uniform system for microRNA annotation". RNA. 9 (3): 277–279. doi:10.1261/rna.2183803. PMC 1370393. PMID 12592000.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Science review of small RNA: Baulcombe D (2002). "DNA events. An RNA microcosm". Science. 297 (5589): 2002–2003. doi:10.1126/science.1077906. PMID 12242426.
- Discovery of lin-4, the first miRNA to be discovered: Lee RC, Feinbaum RL, Ambros V (1993). "The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14". Cell. 75 (5): 843–854. doi:10.1016/0092-8674(93)90529-Y. PMID 8252621.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- The miRBase database
- The miRNA Blog
- miR2Disease, a manually curated database documenting known relationships between miRNA dysregulation and human disease.
- semirna, Web application to search for microRNAs in a plant genome.
- miRandola: Extracellular Circulating microRNAs Database.
