Corynebacterium: Difference between revisions
m WP:CHECKWIKI error fix for #61. Punctuation goes before References. Do general fixes if a problem exists. - |
No edit summary |
||
| Line 57: | Line 57: | ||
'''''Corynebacterium''''' ({{IPAc-en|k|ɔːr|ˈ|aɪ|n|ə|b|æ|k|ˌ|t|ɪər|i|ə|m}}, {{IPAc-en|k|ɔːr|ˈ|ɪ|n|ə|-}}) is a [[genus]] of [[bacteria]] that are [[gram-positive bacteria|gram-positive]] and [[aerobe|aerobic]]. They are [[bacillus (shape)|bacilli]] (rod-shaped), and in some phases of life they are, more particularly, [[club (weapon)|club]]-shaped, which inspired the genus name (''coryneform'' means "club-shaped"). |
'''''Corynebacterium''''' ({{IPAc-en|k|ɔːr|ˈ|aɪ|n|ə|b|æ|k|ˌ|t|ɪər|i|ə|m}}, {{IPAc-en|k|ɔːr|ˈ|ɪ|n|ə|-}}) is a [[genus]] of [[bacteria]] that are [[gram-positive bacteria|gram-positive]] and [[aerobe|aerobic]]. They are [[bacillus (shape)|bacilli]] (rod-shaped), and in some phases of life they are, more particularly, [[club (weapon)|club]]-shaped, which inspired the genus name (''coryneform'' means "club-shaped"). |
||
They are widely distributed in nature in the [[microbiota]] of [[animal]]s (including the [[human microbiota]]) and are mostly innocuous.<ref name="caspium">{{cite journal |
They are widely distributed in nature in the [[microbiota]] of [[animal]]s (including the [[human microbiota]]) and are mostly innocuous.<ref name="caspium">{{cite journal |doi=10.1099/ijs.0.02950-0 }}</ref> Some are useful in industrial settings such as ''C. glutamicum''.<ref name="BurkovskiA">{{cite book |author=Burkovski A (editor). |title=Corynebacteria: Genomics and Molecular Biology |publisher=Caister Academic Press |year=2008 |url=http://www.horizonpress.com/cory |isbn=1-904455-30-1}}{{pn}}</ref> Others can cause human disease, including most notably [[diphtheria]], which is caused by ''[[Corynebacterium diphtheriae|C. diphtheriae]]''. As with various species of a microbiota (including their cousins in the genera ''[[Arcanobacterium]]'' and ''Trueperella''), they usually are not [[pathogen]]ic but can occasionally [[opportunistic infection|opportunistically]] capitalize on atypical access to [[tissue (biology)|tissues]] (via [[wound]]s) or [[immunodeficiency|weakened host defenses]]. |
||
== Taxonomy == |
== Taxonomy == |
||
The genus ''Corynebacterium'' was created by Lehmann and Neumann in 1896 as a [[taxonomy (biology)|taxonomic]] group to contain the bacterial rods responsible for causing diphtheria. The genus was defined based on [[morphology (biology)|morphological]] characteristics. Based on studies of 16S-[[Ribosomal RNA|rRNA]], they have been grouped into the subdivision of gram-positive [[eubacteria]] with high [[Guanine|G]]:[[Cytosine|C]] content, with close phylogenetic relationship to ''[[Arthrobacter]]'', ''[[Mycobacterium]]'', ''[[Nocardia]]'', and ''[[Streptomyces]]''.<ref>{{cite journal |
The genus ''Corynebacterium'' was created by Lehmann and Neumann in 1896 as a [[taxonomy (biology)|taxonomic]] group to contain the bacterial rods responsible for causing diphtheria. The genus was defined based on [[morphology (biology)|morphological]] characteristics. Based on studies of 16S-[[Ribosomal RNA|rRNA]], they have been grouped into the subdivision of gram-positive [[eubacteria]] with high [[Guanine|G]]:[[Cytosine|C]] content, with close phylogenetic relationship to ''[[Arthrobacter]]'', ''[[Mycobacterium]]'', ''[[Nocardia]]'', and ''[[Streptomyces]]''.<ref>{{cite journal |pmid=2439888 }} |
||
</ref> |
</ref> |
||
The term comes from the [[Greek language|Greek]] κορωνη, ''corönë'' ("knotted rod") and βακτηριον, ''bacterion'' ("rod"). The term "diphtheroids" is used to represent corynebacteria that are non[[pathogenic]]; for example, ''[[Corynebacterium diphtheriae|C. diphtheriae]]'' would be excluded |
The term comes from the [[Greek language|Greek]] κορωνη, ''corönë'' ("knotted rod") and βακτηριον, ''bacterion'' ("rod"). The term "diphtheroids" is used to represent corynebacteria that are non[[pathogenic]]; for example, ''[[Corynebacterium diphtheriae|C. diphtheriae]]'' would be excluded.{{fact}} The term diphtheroid comes from Greek διφθερα, ''diphthera''—prepared hide, leather.{{fact}} |
||
==Genomics== |
==Genomics== |
||
Comparative analysis of corynebacterial genomes has led to the identification of several [[conserved signature indels]] which are unique to the genus. Two examples of these conserved signature indels are a two-amino-acid insertion in a conserved region of the enzyme phosphoribose diphosphate:decaprenyl-phosphate phosphoribosyltransferase and a three-amino-acid insertion in [[acetate kinase]], both of which are found only in ''Corynebacterium'' species. Both of these indels serve as [[Genetic marker|molecular markers]] for species of the genus ''Corynebacterium''. Additionally, 16 conserved signature proteins, which are uniquely found in ''Corynebacterium'' species, have been identified. Three of the conserved signature proteins have homologs found in the ''Dietzia'' genus, which is believed to be the closest related genus to ''Corynebacterium''. In phylogenetic trees based on concatenated protein sequences or 16S rRNA, the genus ''Corynebacterium'' forms a distinct clade, within which is a distinct subclade, cluster I. The cluster is made up of the species ''C. diptheriae, C. pseudotuberculosis, C. ulcerans, C. aurimucosum, C. glutamicum,'' and ''C. efficiens''. This cluster is distinguished by several conserved signature indels, such as a two-amino-acid insertion in LepA and a seven- or eight-amino-acid insertions in RpoC. Also, 21 conserved signature proteins are found only in members of cluster I. Another cluster has been proposed, consisting of ''C. jeikeium'' and ''C. urealyticum'', which is supported by the presence of 19 distinct conserved signature proteins which are unique to these two species.<ref>{{ |
Comparative analysis of corynebacterial genomes has led to the identification of several [[conserved signature indels]] which are unique to the genus. Two examples of these conserved signature indels are a two-amino-acid insertion in a conserved region of the enzyme phosphoribose diphosphate:decaprenyl-phosphate phosphoribosyltransferase and a three-amino-acid insertion in [[acetate kinase]], both of which are found only in ''Corynebacterium'' species. Both of these indels serve as [[Genetic marker|molecular markers]] for species of the genus ''Corynebacterium''. Additionally, 16 conserved signature proteins, which are uniquely found in ''Corynebacterium'' species, have been identified. Three of the conserved signature proteins have homologs found in the ''Dietzia'' genus, which is believed to be the closest related genus to ''Corynebacterium''. In phylogenetic trees based on concatenated protein sequences or 16S rRNA, the genus ''Corynebacterium'' forms a distinct clade, within which is a distinct subclade, cluster I. The cluster is made up of the species ''C. diptheriae, C. pseudotuberculosis, C. ulcerans, C. aurimucosum, C. glutamicum,'' and ''C. efficiens''. This cluster is distinguished by several conserved signature indels, such as a two-amino-acid insertion in LepA and a seven- or eight-amino-acid insertions in RpoC. Also, 21 conserved signature proteins are found only in members of cluster I. Another cluster has been proposed, consisting of ''C. jeikeium'' and ''C. urealyticum'', which is supported by the presence of 19 distinct conserved signature proteins which are unique to these two species.<ref>{{cite journal |doi=10.1128/MMBR.05011-11 }}</ref> Corynebateria have a high [[GC content|G+C content]] ranging from 46-74 mol%.<ref>{{cite book |first1=K.A. |last1=Bernard |first2=G. |last2=Funke |chapter=Genus I. Corynebacterium |editor1-first=M. |editor1-last=Goodfellow |editor2-first=P. |editor2-last=Kampfer |editor3-first=H.J. |editor3-last=Busse |editor4-first=M.E. |editor4-last=Trujillo |editor5-first=K. |editor5-last=Suzuki |editor6-first=W. |editor6-last=Ludwig |editor7-first=W.B. |editor7-last=Whitman |title=Bergey's Manual of Systematic Bacteriology |edition=2nd |publisher=Springer |year=2012 |page=245 }}</ref> |
||
== Characteristics == |
== Characteristics == |
||
The principal features of the ''Corynebacterium'' genus were described by Collins and Cummins in 1986.<ref name=collins>Collins |
The principal features of the ''Corynebacterium'' genus were described by Collins and Cummins in 1986.<ref name=collins>{{cite book |last1=Collins |first1=M. D. |last2=Cummins |first2=C. S. |year=1986 |chapter=Genus Corynebacterium Lehmann and Neumann 1896, 350AL |title=Bergey's Manual of Systematic Bacteriology |volume=2 |pages=1266–76 |editor1-first=P. H. A. |editor1-last=Sneath |editor2-first=N. S. |editor2-last=Mair |editor3-first=M. E. |editor3-last=Sharpe |editor4-first=J. G. |editor4-last=Holt |location=Baltimore |publisher=Williams & Wilkins }}</ref> They are gram-positive, [[catalase]]-positive, non[[spore]]-forming, non[[motility|motile]], rod-shaped bacteria that are straight or slightly curved.<ref name=glaucum>{{cite journal |doi=10.1099/ijs.0.02394-0 }}</ref> [[Metachromatic granules]] are usually present representing stored phosphate regions. Their size falls between 2 and 6 [[micrometer|μm]]s in length and 0.5 μm in diameter. The bacteria group together in a characteristic way, which has been described as the form of a "V", "palisades", or "Chinese letters". They may also appear [[ellipse|elliptical]]. They are [[Aerobic organism|aerobic]] or [[Facultative anaerobic organism|facultatively anaerobic]], [[chemoorganotroph]]s. They are [[pleomorphism (microbiology)|pleomorphic]] through their [[Biological life cycle|lifecycles]], they occur in various lengths, and they frequently have thickenings at either end, depending on the surrounding conditions.<ref>{{cite journal |doi=10.1111/j.1365-2672.1977.tb00689.x }}</ref> |
||
=== Cell wall === |
=== Cell wall === |
||
The [[cell wall]] is distinctive, with a predominance of meso[[diaminopimelic acid]] in the [[murein]] wall<ref name=caspium /><ref name=glaucum /> and many repetitions of [[arabinogalactan]], as well as corynemycolic acid (a [[mycolic acid]] with 22 to 26 [[carbon]] atoms), bound by [[disaccharide]] bonds called L-Rha''p''-(1 → 4)--D-GlcNAc-phosphate. These form a complex commonly seen in ''Corynebacterium'' species: the mycolyl-AG–peptidoglican (mAGP).<ref>{{cite journal |
The [[cell wall]] is distinctive, with a predominance of meso[[diaminopimelic acid]] in the [[murein]] wall<ref name=caspium /><ref name=glaucum /> and many repetitions of [[arabinogalactan]], as well as corynemycolic acid (a [[mycolic acid]] with 22 to 26 [[carbon]] atoms), bound by [[disaccharide]] bonds called L-Rha''p''-(1 → 4)--D-GlcNAc-phosphate. These form a complex commonly seen in ''Corynebacterium'' species: the mycolyl-AG–peptidoglican (mAGP).<ref>{{cite journal |doi=10.1093/glycob/cwl066 }}</ref> |
||
=== Culture === |
=== Culture === |
||
| Line 78: | Line 78: | ||
== Habitat == |
== Habitat == |
||
''Corynebacterium'' species occur commonly in nature in the soil, water, plants, and food products.<ref name=caspium /><ref name=glaucum /> The nondiphtheiroid ''Corynebacterium'' species can even be found in the [[mucosa]] and normal [[skin flora]] of humans and animals.<ref name=caspium /><ref name=glaucum /> Unusual habitats, such as the [[preen gland]] of [[birds]] have been recently reported for ''[[Corynebacterium uropygiale]]''.<ref name="Braun"> |
''Corynebacterium'' species occur commonly in nature in the soil, water, plants, and food products.<ref name=caspium /><ref name=glaucum /> The nondiphtheiroid ''Corynebacterium'' species can even be found in the [[mucosa]] and normal [[skin flora]] of humans and animals.<ref name=caspium /><ref name=glaucum /> Unusual habitats, such as the [[preen gland]] of [[birds]] have been recently reported for ''[[Corynebacterium uropygiale]]''.<ref name="Braun">{{cite journal |doi=10.1016/j.syapm.2015.12.001 }}</ref> Some species are known for their pathogenic effects in humans and other animals. Perhaps the most notable one is [[Corynebacterium diphtheriae|''C. diphtheriae'']], which acquires the capacity to produce [[diphtheria toxin]] only after interacting with a [[bacteriophage]].<ref name=costa>{{cite journal |pmid=6270058 }}</ref> Other pathogenic species in humans include: [[Corynebacterium amicolatum|''C. amicolatum'']], [[Corynebacterium striatum|''C. striatum'']], [[Corynebacterium jeikeium|''C. jeikeium'']], [[Corynebacterium urealyticum|''C. urealyticum'']], and [[Corynebacterium xerosis|''C. xerosis'']];<ref>{{cite journal |doi=10.1016/S0213-005X(01)72578-5 }}</ref><ref>{{cite journal |doi=10.1016/S0732-8893(97)00193-4 }}</ref><ref>{{cite journal |doi=10.7547/87507315-85-6-338 }}</ref><ref>{{cite journal |doi=10.1128/aac.23.3.506 }}</ref><ref>{{cite journal |doi=10.1111/j.1574-6968.1983.tb00305.x }}</ref> all of these are important as pathogens in [[immunosuppression|immunosuppressed]] patients. Pathogenic species in other animals include [[Corynebacterium bovis|''C. bovis'']] and [[Corynebacterium renale|''C. renale'']].<ref>{{cite ournal |pmid=8720950 }}</ref> |
||
==Role in disease== |
==Role in disease== |
||
{{main article|Diphtheria}} |
{{main article|Diphtheria}} |
||
The most notable human infection is [[diphtheria]], caused by ''C. diphtheriae''. It is an acute and contagious infection characterized by pseudomembranes of dead [[epithelial]] [[Cell (biology)|cell]]s, [[white blood cell]]s, [[red blood cell]]s, and [[fibrin]] that form around the [[tonsil]]s and [[pharynx|back of the throat]].<ref>[https://www.nlm.nih.gov/medlineplus/spanish/ency/article/001608.htm MedlinePlus - Difteria]</ref> In developed countries, it is an uncommon illness that tends to occur in un[[vaccine|vaccinated]] individuals, especially school-aged children, [[elderly]], [[neutropenia|neutropenic]] or [[immunocompromise]]d patients, and those with prosthetic devices such as [[prosthetic heart valve]]s, [[shunt (medical)|shunts]], or [[catheter]]s. It is more common in developing countries<ref>{{cite journal |doi=10.1590/S0034-89101980000400005 |
The most notable human infection is [[diphtheria]], caused by ''C. diphtheriae''. It is an acute and contagious infection characterized by pseudomembranes of dead [[epithelial]] [[Cell (biology)|cell]]s, [[white blood cell]]s, [[red blood cell]]s, and [[fibrin]] that form around the [[tonsil]]s and [[pharynx|back of the throat]].<ref>[https://www.nlm.nih.gov/medlineplus/spanish/ency/article/001608.htm MedlinePlus - Difteria]</ref> In developed countries, it is an uncommon illness that tends to occur in un[[vaccine|vaccinated]] individuals, especially school-aged children, [[elderly]], [[neutropenia|neutropenic]] or [[immunocompromise]]d patients, and those with prosthetic devices such as [[prosthetic heart valve]]s, [[shunt (medical)|shunts]], or [[catheter]]s. It is more common in developing countries<ref>{{cite journal |doi=10.1590/S0034-89101980000400005 }}</ref> It can occasionally infect wounds, the [[vulva]], the [[conjunctiva]], and the [[middle ear]]. It can be spread [[nosocomial infection|within a hospital]].<ref name=jk>{{cite journal |doi=10.1017/S0022172400065347 }}</ref> The virulent and toxigenic strains are [[Lysogenic cycle|lysogenic]], and produce an [[exotoxin]] formed by two [[polypeptide]] chains, which is itself produced when a bacterium is [[Transformation (genetics)|transformed]] by a [[gene]] from the β [[prophage]].<ref name=costa /> |
||
Several species cause disease in animals, most notably ''C. pseudotuberculosis'', which causes the disease [[caseous lymphadenitis]], and some are also pathogenic in humans. Some attack healthy [[Host (biology)|host]]s, while others tend to attack the [[immunocompromise]]d. Effects of infection include [[granuloma]]tous [[lymphadenopathy]], [[pneumonitis]], [[pharyngitis]], skin infections, and [[endocarditis]]. Corynebacterial endocarditis is seen most frequently in patients with intravascular devices.<ref>{{cite journal |doi=10.1157/13056889 |
Several species cause disease in animals, most notably ''C. pseudotuberculosis'', which causes the disease [[caseous lymphadenitis]], and some are also pathogenic in humans. Some attack healthy [[Host (biology)|host]]s, while others tend to attack the [[immunocompromise]]d. Effects of infection include [[granuloma]]tous [[lymphadenopathy]], [[pneumonitis]], [[pharyngitis]], skin infections, and [[endocarditis]]. Corynebacterial endocarditis is seen most frequently in patients with intravascular devices.<ref>{{cite journal |doi=10.1157/13056889 }}</ref> ''[[Corynebacterium tenuis|C. tenuis]]'' is believed to cause [[trichomycosis palmellina]] and [[trichomycosis axillaris]].<ref>{{EMedicine|derm|601|Trichomycosis axilarris}}</ref> ''C. striatum'' may cause axillary odor.<ref>{{cite journal |doi=10.1111/j.1467-2494.2004.00255.x }}</ref> ''[[Corynebacterium minutissimum|C. minutissimum]]'' causes [[erythrasma]]. |
||
==Industrial uses== |
==Industrial uses== |
||
Nonpathogenic species of ''Corynebacterium'' are used for very important industrial applications, such as the production of [[amino acid]]s,<ref>{{cite book | |
Nonpathogenic species of ''Corynebacterium'' are used for very important industrial applications, such as the production of [[amino acid]]s,<ref>{{cite book |last1=Hongo |first1=M. |last2=Oki |first2=T. |last3=Ogata |first3=S. |chapter=Phage contamination and control |editor1-first=K. |editor1-last=Yamada |editor2-first=S |editor2-last=Kinoshita |editor3-first=T. |editor3-last=Tsunoda |editor4-first=K. |editor4-last=Aida |title=The Microbial Production of Amino Acids |publisher=John Wiley |location=New York |year=1972 |pages=63–83 }}</ref><ref>{{cite book |editor1-last=Yamada |editor1-first=K. |editor2-last=Kinoshita |editor2-first=S. |editor3-last=Tsunoda |editor3-first=T. |editor4-last=Aida |editor4-first=K. |year=1972 |title=The Microbial Production of Amino Acids |publisher=Wiley |location=New York }}{{pn}}</ref> [[nucleotide]]s, and other nutritional factors (Martín, 1989); bioconversion of [[steroid]]s;<ref>{{cite journal |doi=10.1002/bit.260220110 }}</ref> degradation of [[hydrocarbon]]s;<ref>{{cite journal |pmid=422512 }}</ref> [[cheese]] aging;<ref>{{cite journal |doi=10.1111/j.1574-6968.1985.tb01591.x }}</ref> and production of [[enzyme]]s.<ref>Khurana et al., 2000{{full}}</ref> Some species produce metabolites similar to [[antibiotic]]s: [[bacteriocin]]s of the corynecin-linocin type,<ref name=jk /><ref>{{cite journal |doi=10.1111/j.1574-6968.1984.tb01451.x }}</ref><ref>{{cite journal |doi=10.1271/bbb1961.36.2223 }}</ref> antitumor agents,<ref>{{cite journal |doi=10.1016/S0065-230X(08)60090-1 }}</ref> etc. One of the most studied species is [[Corynebacterium glutamicum|''C. glutamicum'']], whose name refers to its capacity to produce [[glutamic acid]] in aerobic conditions.<ref>{{cite journal |doi=10.2323/jgam.13.279 }}</ref> This is used in the food industry as [[monosodium glutamate]] in the production of [[soy sauce]] and [[yogurt]].{{fact}} |
||
Species of ''Corynebacterium'' have been used in the mass production of various amino acids including [[L-Glutamic Acid|glutamic acid]], a food additive that is made at a rate of 1.5 million tons/ year. The metabolic pathways of ''Corynebacterium '' have been further manipulated to produce [[L-Lysine|lysine]] and [[L-threonine|threonine]]. |
Species of ''Corynebacterium'' have been used in the mass production of various amino acids including [[L-Glutamic Acid|glutamic acid]], a food additive that is made at a rate of 1.5 million tons/ year. The metabolic pathways of ''Corynebacterium '' have been further manipulated to produce [[L-Lysine|lysine]] and [[L-threonine|threonine]].{{fact}} |
||
L-Lysine production is specific to ''C. glutamicum'' in which core metabolic enzymes are manipulated through genetic engineering to drive metabolic flux towards the production of NADPH from the pentose phosphate pathway, and L-4-aspartyl phosphate, the commitment step to the synthesis of L-lysine, [[lysC]], dapA, dapC, and dapF. These enzymes are up-regulated in industry through genetic engineering to ensure adequate amounts of lysine precursors are produced to increase metabolic flux. Unwanted side reactions such as threonine and asparagine production can occur if a buildup of intermediates occurs, so scientists have developed mutant strains of'' C. glutamicum'' through PCR engineering and chemical knockouts to ensure production of side-reaction enzymes are limited. Many genetic manipulations conducted in industry are by traditional cross-over methods or inhibition of transcriptional activators |
L-Lysine production is specific to ''C. glutamicum'' in which core metabolic enzymes are manipulated through genetic engineering to drive metabolic flux towards the production of NADPH from the pentose phosphate pathway, and L-4-aspartyl phosphate, the commitment step to the synthesis of L-lysine, [[lysC]], dapA, dapC, and dapF. These enzymes are up-regulated in industry through genetic engineering to ensure adequate amounts of lysine precursors are produced to increase metabolic flux. Unwanted side reactions such as threonine and asparagine production can occur if a buildup of intermediates occurs, so scientists have developed mutant strains of'' C. glutamicum'' through PCR engineering and chemical knockouts to ensure production of side-reaction enzymes are limited. Many genetic manipulations conducted in industry are by traditional cross-over methods or inhibition of transcriptional activators.<ref>{{cite book |last1=Kjeldsen |first1=Kjeld Raunkjær |year=2009 |title=Optimization of an industrial L-lysine producing Corynebacterium glutamicum strain |type=PhD Thesis |publisher=Technical University of Denmark |oclc=826400572 }}{{pn}}</ref> |
||
Expression of functionally active human [[epidermal growth factor]] has been brought about in ''C. glutamicum'',<ref> |
Expression of functionally active human [[epidermal growth factor]] has been brought about in ''C. glutamicum'',<ref>{{cite journal |doi=10.1111/j.1472-765X.2005.01802.x }}</ref> thus demonstrating a potential for industrial-scale production of human proteins. Expressed proteins can be targeted for secretion through either the general [[secretory pathway]] or the [[twin-arginine translocation pathway]].<ref>{{cite journal |doi=10.1007/s00253-007-0934-8 }}</ref> |
||
Unlike gram-negative bacteria, the gram-positive ''Corynebacterium'' species lack [[lipopolysaccharide]]s that function as antigenic [[endotoxins]] in humans. |
Unlike gram-negative bacteria, the gram-positive ''Corynebacterium'' species lack [[lipopolysaccharide]]s that function as antigenic [[endotoxins]] in humans.{{fact}} |
||
==Species== |
==Species== |
||
*''[[Corynebacterium efficiens]]'' <!-- could not work out if it was lipophilic or not --> |
*''[[Corynebacterium efficiens]]'' <!-- could not work out if it was lipophilic or not --> |
||
Most species of corynebacteria are not [[lipophilic bacteria|lipophilic]]. |
Most species of corynebacteria are not [[lipophilic bacteria|lipophilic]].{{fact}} |
||
===Nonlipophilic=== |
===Nonlipophilic=== |
||
| Line 114: | Line 114: | ||
**''[[Corynebacterium matruchotii]]'' |
**''[[Corynebacterium matruchotii]]'' |
||
**''[[Corynebacterium glutamicum]]'' |
**''[[Corynebacterium glutamicum]]'' |
||
**''Corynebacterium'' sp.<ref name=cmr>{{cite journal |pmid=8993861 }}</ref> |
|||
**''Corynebacterium'' sp.<ref name=cmr>Until this point in list the ref is: {{cite journal |vauthors=Funke G, von Graevenitz A, Clarridge JE, Bernard KA |title=Clinical microbiology of coryneform bacteria |journal=Clin. Microbiol. Rev. |volume=10 |issue=1 |pages=125–59 |date=January 1997 |pmid=8993861 |pmc=172946 |url=http://cmr.asm.org/cgi/pmidlookup?view=long&pmid=8993861}}</ref> |
|||
*Nonfermentative corynebacteria |
*Nonfermentative corynebacteria |
||
**''[[Corynebacterium afermentans]]'' subsp. ''afermentans'' |
**''[[Corynebacterium afermentans]]'' subsp. ''afermentans'' |
||
| Line 132: | Line 132: | ||
===Novel corynebacteria that do not contain mycolic acids=== |
===Novel corynebacteria that do not contain mycolic acids=== |
||
*''[[Corynebacterium kroppenstedtii]]''<ref>{{cite journal |
*''[[Corynebacterium kroppenstedtii]]''<ref>{{cite journal |doi=10.1099/00207713-48-4-1449 }}</ref> |
||
==References== |
==References== |
||
| Line 138: | Line 138: | ||
{{Wikispecies|Corynebacterium}} |
{{Wikispecies|Corynebacterium}} |
||
| ⚫ | |||
| ⚫ | |||
* {{cite book | author = Ryan KJ; Ray CG (editors) | title = Sherris Medical Microbiology | edition = 4th | publisher = McGraw Hill | year = 2004 | isbn = 0-8385-8529-9 }} |
* {{cite book | author = Ryan KJ; Ray CG (editors) | title = Sherris Medical Microbiology | edition = 4th | publisher = McGraw Hill | year = 2004 | isbn = 0-8385-8529-9 }} |
||
* [http://www.coryneregnet.de Database of Corynebacterial Transcription Factors and Regulatory Networks] |
* [http://www.coryneregnet.de Database of Corynebacterial Transcription Factors and Regulatory Networks] |
||
* Rollins, David M. University of Maryland: Pathogentic Microbiology: Corynebacterium [http://www.life.umd.edu/classroom/bsci424/PathogenDescriptions/Corynebacterium.htm] |
* Rollins, David M. University of Maryland: Pathogentic Microbiology: Corynebacterium [http://www.life.umd.edu/classroom/bsci424/PathogenDescriptions/Corynebacterium.htm] |
||
| ⚫ | |||
| ⚫ | |||
{{Bacteria classification}} |
{{Bacteria classification}} |
||
Revision as of 15:53, 2 January 2017
| Corynebacterium | |
|---|---|
| |
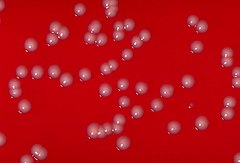
| C. ulcerans colonies on a blood agar plate | |
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Order: | |
| Suborder: | |
| Family: | Corynebacteriaceae
|
| Genus: | Corynebacterium Lehmann & Neumann 1896
|
| Species | |
|
C. accolens | |
Corynebacterium (/kɔːrˈaɪnəbækˌtɪəriəm/, /kɔːrˈɪnə-/) is a genus of bacteria that are gram-positive and aerobic. They are bacilli (rod-shaped), and in some phases of life they are, more particularly, club-shaped, which inspired the genus name (coryneform means "club-shaped").
They are widely distributed in nature in the microbiota of animals (including the human microbiota) and are mostly innocuous.[1] Some are useful in industrial settings such as C. glutamicum.[2] Others can cause human disease, including most notably diphtheria, which is caused by C. diphtheriae. As with various species of a microbiota (including their cousins in the genera Arcanobacterium and Trueperella), they usually are not pathogenic but can occasionally opportunistically capitalize on atypical access to tissues (via wounds) or weakened host defenses.
Taxonomy
The genus Corynebacterium was created by Lehmann and Neumann in 1896 as a taxonomic group to contain the bacterial rods responsible for causing diphtheria. The genus was defined based on morphological characteristics. Based on studies of 16S-rRNA, they have been grouped into the subdivision of gram-positive eubacteria with high G:C content, with close phylogenetic relationship to Arthrobacter, Mycobacterium, Nocardia, and Streptomyces.[3]
The term comes from the Greek κορωνη, corönë ("knotted rod") and βακτηριον, bacterion ("rod"). The term "diphtheroids" is used to represent corynebacteria that are nonpathogenic; for example, C. diphtheriae would be excluded.[citation needed] The term diphtheroid comes from Greek διφθερα, diphthera—prepared hide, leather.[citation needed]
Genomics
Comparative analysis of corynebacterial genomes has led to the identification of several conserved signature indels which are unique to the genus. Two examples of these conserved signature indels are a two-amino-acid insertion in a conserved region of the enzyme phosphoribose diphosphate:decaprenyl-phosphate phosphoribosyltransferase and a three-amino-acid insertion in acetate kinase, both of which are found only in Corynebacterium species. Both of these indels serve as molecular markers for species of the genus Corynebacterium. Additionally, 16 conserved signature proteins, which are uniquely found in Corynebacterium species, have been identified. Three of the conserved signature proteins have homologs found in the Dietzia genus, which is believed to be the closest related genus to Corynebacterium. In phylogenetic trees based on concatenated protein sequences or 16S rRNA, the genus Corynebacterium forms a distinct clade, within which is a distinct subclade, cluster I. The cluster is made up of the species C. diptheriae, C. pseudotuberculosis, C. ulcerans, C. aurimucosum, C. glutamicum, and C. efficiens. This cluster is distinguished by several conserved signature indels, such as a two-amino-acid insertion in LepA and a seven- or eight-amino-acid insertions in RpoC. Also, 21 conserved signature proteins are found only in members of cluster I. Another cluster has been proposed, consisting of C. jeikeium and C. urealyticum, which is supported by the presence of 19 distinct conserved signature proteins which are unique to these two species.[4] Corynebateria have a high G+C content ranging from 46-74 mol%.[5]
Characteristics
The principal features of the Corynebacterium genus were described by Collins and Cummins in 1986.[6] They are gram-positive, catalase-positive, nonspore-forming, nonmotile, rod-shaped bacteria that are straight or slightly curved.[7] Metachromatic granules are usually present representing stored phosphate regions. Their size falls between 2 and 6 μms in length and 0.5 μm in diameter. The bacteria group together in a characteristic way, which has been described as the form of a "V", "palisades", or "Chinese letters". They may also appear elliptical. They are aerobic or facultatively anaerobic, chemoorganotrophs. They are pleomorphic through their lifecycles, they occur in various lengths, and they frequently have thickenings at either end, depending on the surrounding conditions.[8]
Cell wall
The cell wall is distinctive, with a predominance of mesodiaminopimelic acid in the murein wall[1][7] and many repetitions of arabinogalactan, as well as corynemycolic acid (a mycolic acid with 22 to 26 carbon atoms), bound by disaccharide bonds called L-Rhap-(1 → 4)--D-GlcNAc-phosphate. These form a complex commonly seen in Corynebacterium species: the mycolyl-AG–peptidoglican (mAGP).[9]
Culture
Corynebacteria grow slowly, even on enriched media. In terms of nutritional requirements, all need biotin to grow. Some strains also need thiamine and PABA.[6] Some of the Corynebacterium species with sequenced genomes have between 2.5 and 3.0 million base pairs. The bacteria grow in Loeffler's medium, blood agar, and trypticase soy agar (TSA). They form small, grayish colonies with a granular appearance, mostly translucent, but with opaque centers, convex, with continuous borders.[7] The color tends to be yellowish-white in Loeffler's medium. In TSA, they can form grey colonies with black centers and dentated borders that look similar to flowers (C. gravis), or continuous borders (C. mitis), or a mix between the two forms (C. intermedium).
Habitat
Corynebacterium species occur commonly in nature in the soil, water, plants, and food products.[1][7] The nondiphtheiroid Corynebacterium species can even be found in the mucosa and normal skin flora of humans and animals.[1][7] Unusual habitats, such as the preen gland of birds have been recently reported for Corynebacterium uropygiale.[10] Some species are known for their pathogenic effects in humans and other animals. Perhaps the most notable one is C. diphtheriae, which acquires the capacity to produce diphtheria toxin only after interacting with a bacteriophage.[11] Other pathogenic species in humans include: C. amicolatum, C. striatum, C. jeikeium, C. urealyticum, and C. xerosis;[12][13][14][15][16] all of these are important as pathogens in immunosuppressed patients. Pathogenic species in other animals include C. bovis and C. renale.[17]
Role in disease
The most notable human infection is diphtheria, caused by C. diphtheriae. It is an acute and contagious infection characterized by pseudomembranes of dead epithelial cells, white blood cells, red blood cells, and fibrin that form around the tonsils and back of the throat.[18] In developed countries, it is an uncommon illness that tends to occur in unvaccinated individuals, especially school-aged children, elderly, neutropenic or immunocompromised patients, and those with prosthetic devices such as prosthetic heart valves, shunts, or catheters. It is more common in developing countries[19] It can occasionally infect wounds, the vulva, the conjunctiva, and the middle ear. It can be spread within a hospital.[20] The virulent and toxigenic strains are lysogenic, and produce an exotoxin formed by two polypeptide chains, which is itself produced when a bacterium is transformed by a gene from the β prophage.[11]
Several species cause disease in animals, most notably C. pseudotuberculosis, which causes the disease caseous lymphadenitis, and some are also pathogenic in humans. Some attack healthy hosts, while others tend to attack the immunocompromised. Effects of infection include granulomatous lymphadenopathy, pneumonitis, pharyngitis, skin infections, and endocarditis. Corynebacterial endocarditis is seen most frequently in patients with intravascular devices.[21] C. tenuis is believed to cause trichomycosis palmellina and trichomycosis axillaris.[22] C. striatum may cause axillary odor.[23] C. minutissimum causes erythrasma.
Industrial uses
Nonpathogenic species of Corynebacterium are used for very important industrial applications, such as the production of amino acids,[24][25] nucleotides, and other nutritional factors (Martín, 1989); bioconversion of steroids;[26] degradation of hydrocarbons;[27] cheese aging;[28] and production of enzymes.[29] Some species produce metabolites similar to antibiotics: bacteriocins of the corynecin-linocin type,[20][30][31] antitumor agents,[32] etc. One of the most studied species is C. glutamicum, whose name refers to its capacity to produce glutamic acid in aerobic conditions.[33] This is used in the food industry as monosodium glutamate in the production of soy sauce and yogurt.[citation needed]
Species of Corynebacterium have been used in the mass production of various amino acids including glutamic acid, a food additive that is made at a rate of 1.5 million tons/ year. The metabolic pathways of Corynebacterium have been further manipulated to produce lysine and threonine.[citation needed]
L-Lysine production is specific to C. glutamicum in which core metabolic enzymes are manipulated through genetic engineering to drive metabolic flux towards the production of NADPH from the pentose phosphate pathway, and L-4-aspartyl phosphate, the commitment step to the synthesis of L-lysine, lysC, dapA, dapC, and dapF. These enzymes are up-regulated in industry through genetic engineering to ensure adequate amounts of lysine precursors are produced to increase metabolic flux. Unwanted side reactions such as threonine and asparagine production can occur if a buildup of intermediates occurs, so scientists have developed mutant strains of C. glutamicum through PCR engineering and chemical knockouts to ensure production of side-reaction enzymes are limited. Many genetic manipulations conducted in industry are by traditional cross-over methods or inhibition of transcriptional activators.[34]
Expression of functionally active human epidermal growth factor has been brought about in C. glutamicum,[35] thus demonstrating a potential for industrial-scale production of human proteins. Expressed proteins can be targeted for secretion through either the general secretory pathway or the twin-arginine translocation pathway.[36]
Unlike gram-negative bacteria, the gram-positive Corynebacterium species lack lipopolysaccharides that function as antigenic endotoxins in humans.[citation needed]
Species
Most species of corynebacteria are not lipophilic.[citation needed]
Nonlipophilic
The nonlipophilic bacteria may be classified as fermentative and nonfermentative:
- Fermentative corynebacteria
- Nonfermentative corynebacteria
Lipophilic
- Corynebacterium uropygiale[10]
- Corynebacterium jeikeium
- Corynebacterium urealyticum
- Corynebacterium afermentans subsp. lipophilum
- Corynebacterium accolens
- Corynebacterium macginleyi
- CDC coryneform groups F-1 and G
- Corynebacterium bovis[37]
Novel corynebacteria that do not contain mycolic acids
References
- ^ a b c d . doi:10.1099/ijs.0.02950-0.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Burkovski A (editor). (2008). Corynebacteria: Genomics and Molecular Biology. Caister Academic Press. ISBN 1-904455-30-1.
{{cite book}}:|author=has generic name (help)[page needed] - ^ . PMID 2439888.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1128/MMBR.05011-11.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Bernard, K.A.; Funke, G. (2012). "Genus I. Corynebacterium". In Goodfellow, M.; Kampfer, P.; Busse, H.J.; Trujillo, M.E.; Suzuki, K.; Ludwig, W.; Whitman, W.B. (eds.). Bergey's Manual of Systematic Bacteriology (2nd ed.). Springer. p. 245.
- ^ a b Collins, M. D.; Cummins, C. S. (1986). "Genus Corynebacterium Lehmann and Neumann 1896, 350AL". In Sneath, P. H. A.; Mair, N. S.; Sharpe, M. E.; Holt, J. G. (eds.). Bergey's Manual of Systematic Bacteriology. Vol. 2. Baltimore: Williams & Wilkins. pp. 1266–76.
- ^ a b c d e . doi:10.1099/ijs.0.02394-0.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1111/j.1365-2672.1977.tb00689.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1093/glycob/cwl066.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ a b . doi:10.1016/j.syapm.2015.12.001.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ a b . PMID 6270058.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1016/S0213-005X(01)72578-5.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1016/S0732-8893(97)00193-4.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.7547/87507315-85-6-338.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1128/aac.23.3.506.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1111/j.1574-6968.1983.tb00305.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Template:Cite ournal
- ^ MedlinePlus - Difteria
- ^ . doi:10.1590/S0034-89101980000400005.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ a b . doi:10.1017/S0022172400065347.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1157/13056889.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Trichomycosis axilarris at eMedicine
- ^ . doi:10.1111/j.1467-2494.2004.00255.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Hongo, M.; Oki, T.; Ogata, S. (1972). "Phage contamination and control". In Yamada, K.; Kinoshita, S; Tsunoda, T.; Aida, K. (eds.). The Microbial Production of Amino Acids. New York: John Wiley. pp. 63–83.
- ^ Yamada, K.; Kinoshita, S.; Tsunoda, T.; Aida, K., eds. (1972). The Microbial Production of Amino Acids. New York: Wiley.[page needed]
- ^ . doi:10.1002/bit.260220110.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . PMID 422512.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1111/j.1574-6968.1985.tb01591.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Khurana et al., 2000[full citation needed]
- ^ . doi:10.1111/j.1574-6968.1984.tb01451.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1271/bbb1961.36.2223.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1016/S0065-230X(08)60090-1.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.2323/jgam.13.279.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ Kjeldsen, Kjeld Raunkjær (2009). Optimization of an industrial L-lysine producing Corynebacterium glutamicum strain (PhD Thesis). Technical University of Denmark. OCLC 826400572.[page needed]
- ^ . doi:10.1111/j.1472-765X.2005.01802.x.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1007/s00253-007-0934-8.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ a b c . PMID 8993861.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help) - ^ . doi:10.1099/00207713-48-4-1449.
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help)
Further reading
- Burkovski, Andreas, ed. (2008). Corynebacteria: Genomics and Molecular Biology. Caister Academic Press. ISBN 1-904455-30-1.
- Ryan KJ; Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 0-8385-8529-9.
{{cite book}}:|author=has generic name (help)CS1 maint: multiple names: authors list (link) - Database of Corynebacterial Transcription Factors and Regulatory Networks
- Rollins, David M. University of Maryland: Pathogentic Microbiology: Corynebacterium [1]
