SH3 domain: Difference between revisions
m WP:CHECKWIKI error fixes + genfixes using AWB (7117) |
→Proteins with SH3 domain: added HIV integrase |
||
| Line 40: | Line 40: | ||
*[[GRB2]] |
*[[GRB2]] |
||
*[[p54 S6 kinase 2 (S6K2)]] |
*[[p54 S6 kinase 2 (S6K2)]] |
||
*[[Integrase|HIV Integrase]] |
|||
==See also== |
==See also== |
||
Revision as of 02:38, 19 April 2011
| SH3 domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
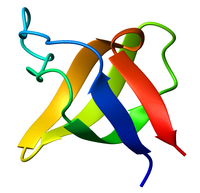 Ribbon diagram of the SH3 domain, alpha spectrin, from chicken (PDB accession code 1SHG), colored from blue (N-terminus) to red (C-terminus). | |||||||||
| Identifiers | |||||||||
| Symbol | SH3_1 | ||||||||
| Pfam | PF00018 | ||||||||
| Pfam clan | CL0010 | ||||||||
| InterPro | IPR001452 | ||||||||
| SMART | SM00326 | ||||||||
| PROSITE | PS50002 | ||||||||
| SCOP2 | 1shf / SCOPe / SUPFAM | ||||||||
| |||||||||
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acids residues first identified as a conserved sequence in the viral adaptor protein v-Crk and the non-catalytic parts of enzymes such as phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src.[1][2] It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase activating protein, CDC24 and CDC25.[3][4][5]
The SH3 domain has a characteristic beta-barrel fold which consists of five or six β-strands arranged as two tightly packed anti-parallel β sheets. The linker regions may contain short helices. Approximately 300 SH3 domains are encoded in the human genome. The SH3-type fold is an ancient fold found in eukaryotes as well as prokaryotes.[6] Examples for similar folds are the OB fold, which can, for example, bind to sugars or the HIV DNA Integrase DNA binding domain. Such functionally distinct SH3-like domains often have no recognizable homology on the amino acid level. The classical SH3 domain is usually found in proteins that interact with other proteins and mediate assembly of specific protein complexes, typically via binding to proline-rich peptides in their respective binding partner. Classical SH3 domains are restricted in humans to intracellular proteins, although the small human MIA family of extracellular proteins also contain a domain with an SH3-like fold.
Many SH3-binding epitopes of proteins have a consensus sequence:
-X-P-p-X-P- 1 2 3 4 5
with 1 and 4 being aliphatic amino acids, 2 and 5 always and 3 sometimes being proline. The sequence binds to the hydrophobic pocket of the SH3 domain. More recently, SH3 domains that bind to a core consensus motif R-x-x-K have been described. Examples are the C-terminal SH3 domains of adaptor proteins like Grb2 and Mona (a.k.a. Gads, Grap2, Grf40, GrpL etc.). Other SH3 binding motifs have emerged and are still emerging in the course of various molecular studies, highlighting the versatility of this domain.
SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. They also regulate the activity state of adaptor proteins and other tyrosine kinases and are thought to increase the substrate specificity of some tyrosine kinases by binding far away from the catalytic center of the kinase.
Proteins with SH3 domain
- Adaptor proteins
- CDC24
- CDC25
- PI3 kinase
- Phospholipase
- Ras GTPase activating protein
- Vav proto-oncogene
- GRB2
- p54 S6 kinase 2 (S6K2)
- HIV Integrase
See also
References
- ^ Pawson T, Schlessingert J (1993). "SH2 and SH3 domains". Curr. Biol. 3 (7): 434–42. PMID 15335710.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Mayer BJ (2001). "SH3 domains: complexity in moderation". J. Cell. Sci. 114 (Pt 7): 1253–63. PMID 11256992.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Musacchio A, Gibson T, Lehto VP, Saraste M (1992). "SH3--an abundant protein domain in search of a function". FEBS Lett. 307 (1): 55–61. PMID 1639195.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Mayer BJ, Baltimore D (1993). "Signalling through SH2 and SH3 domains". Trends Cell Biol. 3 (1): 8–13. PMID 14731533.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Pawson T (1995). "Protein modules and signalling networks". Nature. 373 (6515): 573–80. doi:10.1038/373573a0. PMID 7531822.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Whisstock JC, Lesk AM (1999). "SH3 domains in prokaryotes". Trends Biochem. Sci. 24 (4): 132–3. PMID 10322416.
{{cite journal}}: Unknown parameter|month=ignored (help)
External links
- Nash Lab Protein Interaction Domains in Signal Transduction - The SH3 domain
- GENEART - Screen your protein against all human SH3 domains in a single phage display cycle
