Telomere
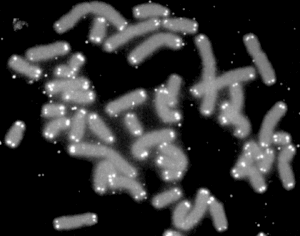
A telomere is a region of repetitive DNA sequences at the end of a chromosome, which protects the end of the chromosome from deterioration or from fusion with neighbouring chromosomes. Its name is derived from the Greek nouns telos (τέλος) "end" and merοs (μέρος, root: μερ-) "part". The telomere regions deter the degradation of genes near the ends of chromosomes by allowing for the shortening of chromosome ends, which necessarily occurs during chromosome replication.[1] Over time, due to each cell division, the telomere ends do become shorter.[2]
During cell division, enzymes that duplicate DNA cannot continue their duplication all the way to the end of chromosomes. If cells divided without telomeres, they would lose the ends of their chromosomes, and the necessary information they contain. The telomeres are disposable buffers blocking the ends of the chromosomes, are consumed during cell division, and are replenished by an enzyme, telomerase reverse transcriptase.
Background
All living things (organisms) are made up of cells. See also cell (biology) and cell theory. Cells are small structures that give an organism its shape, carry on cellular respiration (consume oxygen and chemicals, release energy, and do work), reproduce, and grow. Every cell contains a DNA chemical "blueprint" giving information on how to make new copies of the cell. This information is stored in the cell's chromosomes and genes. See Introduction to genetics.
Discovery
In the early 1970s, Russian theorist Alexei Olovnikov first recognized the problem of how chromosomes could not completely replicate their ends. Building on this, and to accommodate Leonard Hayflick's idea of limited somatic cell division, Olovnikov suggested that DNA sequences are lost every time a cell/DNA replicates until the loss reaches a critical level, at which point cell division ends.[3][4]
In 1975–1977, Elizabeth Blackburn, working as a postdoctoral fellow at Yale University with Joseph Gall, discovered the unusual nature of telomeres, with their simple repeated DNA sequences composing chromosome ends. Their work was published in 1978. The telomere shortening mechanism normally limits cells to a fixed number of divisions, and animal studies suggest that this is responsible for aging on the cellular level and sets a limit on lifespans. Telomeres protect a cell's chromosomes from fusing with each other or rearranging — abnormalities that can lead to cancer — and so cells are destroyed when their telomeres are consumed. Most cancers are the result of "immortal" cells that have ways of evading this programmed destruction.[5]
Elizabeth Blackburn, Carol Greider, and Jack Szostak were awarded the 2009 Nobel Prize in Physiology or Medicine for the discovery of how chromosomes are protected by telomeres and the enzyme telomerase.[6]
Nature and function of telomeres
Structure, function and evolutionary biology
Telomeres are repetitive DNA sequences located at the termini of linear chromosomes of most eukaryotic organisms. Because most prokaryotes have circular chromosomes, the majority of them do not have telomeres. Telomeres compensate for incomplete semi-conservative DNA replication at chromosomal ends. The protection against homologous recombination (HR) and non-homologous end joining (NHEJ) constitutes the essential “capping” role of telomeres that distinguishes them from DNA double-strand breaks (DSBs).[7]

In most prokaryotes, chromosomes are circular and, thus, do not have ends to suffer premature replication termination. A small fraction of bacterial chromosomes (such as those in Streptomyces and Borrelia) are linear and possess telomeres, which are very different from those of the eukaryotic chromosomes in structure and functions. The known structures of bacterial telomeres take the form of proteins bound to the ends of linear chromosomes, or hairpin loops of single-stranded DNA at the ends of the linear chromosomes.[8]
While replicating of DNA, the eukaryotic DNA replication enzymes (the DNA polymerase protein complex) cannot replicate the sequences present at the ends of the chromosomes (or more precisely the chromatid fibres). Hence these sequences and the information they carry may get lost. This is the reason why telomeres are so important in context of successful cell division, because they "cap" the end-sequences and themselves get lost in the process of DNA replication. But the cell has an enzyme called telomerase which carries out the task of adding repetitive nucleotide sequences to the ends of the DNA. Telomerase thus "replenishes" the telomere "cap" of the DNA. In most multicellular eukaryotic organisms, telomerase is active only in germ cells, stem cells and certain white blood cells. There are theories that claim that the steady shortening of telomeres with each replication in somatic (body) cells may have a role in senescence and in the prevention of cancer. This is because the telomeres act as a sort of time-delay "fuse", eventually running out after a certain number of cell divisions and resulting in the eventual loss of vital genetic information from the cell's chromosome with future divisions.
Telomere length varies greatly between species, from approximately 300 base pairs in yeast[9] to many kilobases in humans, and usually is composed of arrays of guanine-rich, six- to eight-base-pair-long repeats. Eukaryotic telomeres normally terminate with 3′ single-stranded-DNA overhang, which is essential for telomere maintenance and capping. Multiple proteins binding single- and double-stranded telomere DNA have been identified.[10] These function in both telomere maintenance and capping. Telomeres form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins.[11] At the very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop.[12]
Telomere shortening in humans can induce replicative senescence, which blocks cell division. This mechanism appears to prevent genomic instability and development of cancer in human aged cells by limiting the number of cell divisions. However, shortened telomeres impair immune function which might also increase cancer susceptibility.[13] Malignant cells that bypass this arrest become immortalized by telomere extension due mostly to the activation of telomerase, the reverse transcriptase enzyme responsible for synthesis of telomeres. However, 5–10% of human cancers activate the Alternative Lengthening of Telomeres (ALT) pathway, which relies on recombination-mediated elongation. [citation needed]
Since shorter telomeres are thought to be a cause of poorer health and aging, this raises the question of why longer telomeres are not selected for to ameliorate these effects. A prominent explanation suggests that inheriting longer telomeres would cause increased cancer rates (e.g. Weinstein and Ciszek, 2002). However, a recent literature review and analysis [13] suggests this is unlikely, because shorter telomeres and telomerase inactivation is more often associated with increased cancer rates, and the mortality from cancer occurs late in life when the force of natural selection is very low. An alternative explanation to the hypothesis that long telomeres are selected against due to their cancer promoting effects is the "thrifty telomere" hypothesis which suggests that the cellular proliferation effects of longer telomeres causes increased energy expenditures.[13] In environments of energetic limitation, shorter telomeres might be an energy sparing mechanism.
Human telomeres, cancer and ALT (Alternative lengthening of telomeres)
This section may be too technical for most readers to understand. (December 2010) |
Human somatic cells without telomerase gradually lose telomeric sequences as a result of incomplete replication (Counter et al., 1992). As human telomeres grow shorter, eventually cells reach the limit of their replicative capacity and progress into senescence or old age. Senescence involves p53 and pRb pathways and leads to the halting of cell proliferation (Campisi, 2005). Senescence may play an important role in suppression of cancer emergence, although inheriting shorter telomeres probably does not protect against cancer.[13] With critically shortened telomeres, further cell proliferation can be achieved by inactivation of p53 and pRb pathways. Cells entering proliferation after inactivation of p53 and pRb pathways undergo crisis. Crisis is characterized by gross chromosomal rearrangements and genome instability, and almost all cells die. Rare cells emerge from crisis immortalized through telomere lengthening by either activated telomerase or ALT (Colgina and Reddel, 1999; Reddel and Bryan, 2003). The first description of an ALT cell line demonstrated that their telomeres are highly heterogeneous in length and predicted a mechanism involving recombination (Murnane et al., 1994). Subsequent studies have confirmed a role for recombination in telomere maintenance by ALT (Dunham et al., 2000), however the exact mechanism of this pathway is yet to be determined. ALT cells produce abundant t-circles, possible products of intratelomeric recombination and t-loop resolution (Tomaska et al., 2000; 2009; Cesare and Griffith, 2004; Wang et al., 2004).
Telomerase is a "ribonucleoprotein complex" composed of a protein component and an RNA primer sequence that acts to protect the terminal ends of chromosomes. The actions of telomerase are necessary because, during replication, DNA polymerase can synthesize DNA in only a 5' to 3' direction and can do so only by adding polynucleotides to an RNA primer that has already been placed at various points along the length of the DNA. These RNA strands must later be replaced with DNA. This replacement of the RNA primers is not a problem at origins of replication within the chromosome because DNA polymerase can use a previous stretch of DNA 5' to the RNA template as a template to backfill the sequence where the RNA primer was; at the terminal end of the chromosome, however, DNA polymerase cannot replace the RNA primer because there is no position 5' of the RNA primer where another primer can be placed, nor is there DNA upstream that can be used as a primer so that DNA polymerase can replace the RNA primer. Without telomeres at the end of DNA, this genetic sequence at the end of the chromosome would be deleted. The chromosome would grow shorter and shorter in subsequent replications and genetic information would be lost. The telomere prevents this problem by employing a different mechanism to synthesize DNA at this point, thereby preserving the sequence at the terminal of the chromosome. This prevents chromosomal fraying and prevents the ends of the chromosome from being processed as a double-strand DNA break, which could lead to chromosome-to-chromosome telomere fusions. Telomeres are extended by telomerases, part of a protein subgroup of specialized reverse transcriptase enzymes known as TERT (TElomerase Reverse Transcriptases) that are involved in synthesis of telomeres in humans and many other, but not all, organisms. However, because of DNA replication mechanisms, oxidative stress, and, because TERT expression is very low in many types of human cells, the telomeres of these cells shrink a little bit every time a cell divides, although, in other cellular compartments that require extensive cell division, such as stem cells and certain white blood cells, TERT is expressed at higher levels and telomere shortening is partially or fully prevented.

In addition to its TERT protein component, telomerase also contains a piece of template RNA known as the TERC (TElomerase RNA Component) or TR (Telomerase RNA). In humans, this TERC telomere sequence is a repeating string of TTAGGG, between 3 and 20 kilobases in length. There are an additional 100-300 kilobases of telomere-associated repeats between the telomere and the rest of the chromosome. Telomere sequences vary from species to species, but, in general, one strand is rich in G with fewer Cs. These G-rich sequences can form four-stranded structures (G-quadruplexes), with sets of four bases held in plane and then stacked on top of each other with either a sodium or a potassium ion between the planar quadruplexes.
If telomeres become too short, they have the potential to unfold from their presumed closed structure. The cell may detect this uncapping as DNA damage and then either stop growing, enter cellular old age (senescence), or begin programmed cell self-destruction (apoptosis) depending on the cell's genetic background (p53 status). Uncapped telomeres also result in chromosomal fusions. Since this damage cannot be repaired in normal somatic cells, the cell may even go into apoptosis. Many aging-related diseases are linked to shortened telomeres. Organs deteriorate as more and more of their cells die off or enter cellular senescence.
At the very distal end of the telomere is a 300 bp single-stranded portion, which forms the T-Loop. This loop is analogous to a knot, which stabilizes the telomere, preventing the telomere ends from being recognized as break points by the DNA repair machinery. Should non-homologous end joining occur at the telomeric ends, chromosomal fusion will result. The T-loop is held together by seven known proteins, the most notable ones being TRF1, TRF2, POT1, TIN1, and TIN2, collectively referred to as the shelterin complex.
Telomere shortening

Telomeres shorten in part because of the end replication problem that is exhibited during DNA replication in eukaryotes only. Because DNA replication does not begin at either end of the DNA strand, but starts in the center, and considering that all known DNA polymerases move in the 5' to 3' direction, one finds a leading and a lagging strand on the DNA molecule being replicated.
On the leading strand, DNA polymerase can make a complementary DNA strand without any difficulty because it goes from 5' to 3'. However, there is a problem going in the other direction on the lagging strand. To counter this, short sequences of RNA acting as primers attach to the lagging strand a short distance ahead of where the initiation site was. The DNA polymerase can start replication at that point and go to the end of the initiation site. This causes the formation of Okazaki fragments. More RNA primers attach further on the DNA strand and DNA polymerase comes along and continues to make a new DNA strand.
Eventually, the last RNA primer attaches, and DNA polymerase, RNA nuclease, and DNA ligase come along to convert the RNA (of the primers) to DNA and to seal the gaps in between the Okazaki fragments. But, in order to change RNA to DNA, there must be another DNA strand in front of the RNA primer. This happens at all the sites of the lagging strand, but it does not happen at the end where the last RNA primer is attached. Ultimately, that RNA is destroyed by enzymes that degrade any RNA left on the DNA. Thus, a section of the telomere is lost during each cycle of replication at the 5' end of the lagging strand.
However, in vitro studies (von Zglinicki et al. 1995, 2000) have shown that telomeres are highly susceptible to oxidative stress. Telomere shortening due to free radicals explains the difference between the estimated loss per division because of the end-replication problem (ca. 20 bp) and actual telomere shortening rates (50-100 bp), and has a greater absolute impact on telomere length than shortening caused by the end-replication problem.
Lengthening telomeres
The phenomenon of limited cellular division was first observed by Leonard Hayflick, and is now referred to as the Hayflick Limit. Significant discoveries were made by the team led by Professor Elizabeth Blackburn at the University of California, San Francisco (UCSF).
Advocates of human life extension promote the idea of lengthening the telomeres in certain cells through temporary activation of telomerase (by drugs), or possibly permanently by gene therapy. They reason that this would extend human life because it would extend the Hayflick Limit. So far these ideas have not been proven in humans, but it has been demonstrated that telomere extension has successfully reversed some signs of aging in laboratory mice [14][15] and the nematode worm species Caenorhabditis elegans.[16] However, it has been hypothesized that longer telomeres and especially telomerase activation might cause increased cancer (e.g. Weinstein and Ciszek, 2002). Paradoxically, longer telomeres might also protect against cancer, because short telomeres are associated with cancer. It has also been suggested that longer telomeres might cause increased energy consumption.[13]
Techniques to extend telomeres could be useful for tissue engineering, because they might permit healthy, noncancerous mammalian cells to be cultured in amounts large enough to be engineering materials for biomedical repairs.
However, there are several issues that still need to be cleared up. First, it is not even certain whether the relationship between telomeres and aging is causal. Changing telomere lengths are usually associated with changing speed of senescence. This telomere shortening, however, might be a consequence of, and not a reason for, aging.
That the role of telomeres is far from being understood is demonstrated by two recent studies on long-lived seabirds. In 2003, scientists observed that the telomeres of Leach's Storm-petrel (Oceanodroma leucorhoa) seem to lengthen with chronological age, the first observed instance of such behaviour of telomeres.[17] In 2006, Juola et al.[18] reported that in another unrelated, long-lived seabird species, the Great Frigatebird (Fregata minor), telomere length did decrease until at least c.40 years of age (i.e. probably over the entire lifespan), but the speed of decrease slowed down massively with increasing ages, and that rates of telomere length decrease varied strongly between individual birds. They concluded that in this species (and probably in frigatebirds and their relatives in general), telomere length could not be used to determine a bird's age sufficiently well. Thus, it seems that there is much more variation in the behavior of telomere length than initially believed.
The telomere length varies in cloned animals. Sometimes the clones end up with shorter telomeres since the DNA has already divided countless times. Occasionally, the telomeres in a clone's DNA are longer because they get "reprogrammed" [citation needed].
Telomere sequences
| Group | Organism | Telomeric repeat (5' to 3' toward the end) |
|---|---|---|
| Vertebrates | Human, mouse, Xenopus | TTAGGG |
| Filamentous fungi | Neurospora crassa | TTAGGG |
| Slime moulds | Physarum, Didymium | TTAGGG |
| Dictyostelium | AG(1-8) | |
| Kinetoplastid protozoa | Trypanosoma, Crithidia | TTAGGG |
| Ciliate protozoa | Tetrahymena, Glaucoma | TTGGGG |
| Paramecium | TTGGG(T/G) | |
| Oxytricha, Stylonychia, Euplotes | TTTTGGGG | |
| Apicomplexan protozoa | Plasmodium | TTAGGG(T/C) |
| Higher plants | Arabidopsis thaliana | TTTAGGG |
| Green algae | Chlamydomonas | TTTTAGGG |
| Insects | Bombyx mori | TTAGG |
| Roundworms | Ascaris lumbricoides | TTAGGC |
| Fission yeasts | Schizosaccharomyces pombe | TTAC(A)(C)G(1-8) |
| Budding yeasts | Saccharomyces cerevisiae | TGTGGGTGTGGTG (from RNA template) or G(2-3)(TG)(1-6)T (consensus) |
| Saccharomyces castellii | TCTGGGTG | |
| Candida glabrata | GGGGTCTGGGTGCTG | |
| Candida albicans | GGTGTACGGATGTCTAACTTCTT | |
| Candida tropicalis | GGTGTA[C/A]GGATGTCACGATCATT | |
| Candida maltosa | GGTGTACGGATGCAGACTCGCTT | |
| Candida guillermondii | GGTGTAC | |
| Candida pseudotropicalis | GGTGTACGGATTTGATTAGTTATGT | |
| Kluyveromyces lactis | GGTGTACGGATTTGATTAGGTATGT |
Telomeres and cancer
This section needs expansion. You can help by adding to it. (June 2008) |
As a cell begins to become cancerous, it divides more often and its telomeres become very short[citation needed]. If its telomeres get too short, the cell may die. It can escape this fate by up-regulating an enzyme called telomerase, which can prevent telomeres from getting shorter and even elongate them.
Studies have found shortened telomeres in many cancers, including pancreatic, bone, prostate, bladder, lung, kidney, and head and neck. In addition, people with many types of cancer have been found to possess shorter leukocyte telomeres than healthy controls.[19]
Cancer cells require a mechanism to maintain their telomeric DNA in order to continue dividing indefinitely (immortalization). A mechanism for telomere elongation or maintenance is one of the key steps in cellular immortalization and can be used as a diagnostic marker in the clinic. Telomerase, the enzyme complex responsible for elongating telomeres, is activated in approximately 90% of tumors. However, a sizeable fraction of cancerous cells employ alternative lengthening of telomeres (ALT),[20] a non-conservative telomere lengthening pathway involving the transfer of telomere tandem repeats between sister-chromatids.[21]
Telomerase is the natural enzyme that promotes telomere repair. It is active in stem cells, germ cells, hair follicles, and 90 percent of cancer cells, but its expression is low or absent in somatic cells. Telomerase functions by adding bases to the ends of the telomeres. Cells with sufficient telomerase activity are considered immortal in the sense that they can divide past the Hayflick limit without entering senescence or apoptosis. For this reason, telomerase is viewed as a potential target for anti-cancer drugs (such as telomestatin)..[22]
Studies using knockout mice have demonstrated that the role of telomeres in cancer can both be limiting to tumor growth, as well as promote tumorigenesis, depending on the cell type and genomic context.[23][24]
Measurement of telomere length in the laboratory
This section may be too technical for most readers to understand. (June 2009) |
Several techniques are currently employed to assess average telomere length in eukaryotic cells. The most widely used method is the Terminal Restriction Fragment (TRF) southern blot,[25] which involves hybridization of a radioactive 32P-(TTAGGG)n oligonucleotide probe to Hinf / Rsa I digested genomic DNA embedded on a nylon membrane and subsequently exposed to autoradiographic film or phosphoimager screen. Another histochemical method, termed Q-FISH, involves fluorescent in situ hybridization (FISH).[26] Q-FISH, however, requires significant amounts of genomic DNA (2-20 micrograms) and labor that renders its use limited in large epidemiological studies. Some of these impediments have been overcome with a Real-Time PCR assay for telomere length and Flow-FISH. RT-PCR assay involves determining the Telomere-to-Single Copy Gene (T/S)ratio,[27] which is demonstrated to be proportional to the average telomere length in a cell.
Another technique, referred to as single telomere elongation length analysis (STELA), was developed in 2003 by Duncan Baird. This technique allows investigations can target specific telomere ends, which is not possible with TRF analysis. However, due to this technique's being PCR-based, telomeres larger than 25Kb cannot be amplified and there is a bias towards shorter telomeres.
See also
- Senescence (Biological aging).
- Rejuvenation (aging)
- Immortality
References
- ^ AtGoogleTalks, August 20, 2008 Molecular biologist Elizabeth Blackburn
- ^ Passarge, Eberhard. Color atlas of genetics, 2007.
- ^ Olovnikov, Alexei M. (1971). "Принцип маргинотомии в матричном синтезе полинуклеотидов". Doklady Akademii Nauk SSSR (in Russian). 201 (6): 1496–9. PMID 5158754.
{{cite journal}}: Unknown parameter|trans_title=ignored (|trans-title=suggested) (help) - ^ Olovnikov AM (1973). "A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon". J. Theor. Biol. 41 (1): 181–90. doi:10.1016/0022-5193(73)90198-7. PMID 4754905.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Harrison's Principles of Internal Medicine, Ch. 69, Cancer cell biology and angiogenesis, Robert G. Fenton and Dan L. Longo, p. 454.
- ^ http://nobelprize.org/nobel_prizes/medicine/laureates/2009/press.html
- ^ Lundblad, 2000; Ferreira et al., 2004
- ^ Maloy, Stanley (July 12, 2002). "Bacterial Chromosome Structure". Retrieved 2008-06-22.
- ^ Shampay.J., Szostak,J.W. and Blackburn,E.H. (1984) DNA sequences of telomeres maintained in yeast. Nature, 310, 154-157
- ^ Blackburn, 2001; Smogorzewska and de Lange, 2004; Cech, 2004; De Lange et al., 2005; Kota and Runge, 1999
- ^ Griffith J, Comeau L, Rosenfield S, Stansel R, Bianchi A, Moss H, de Lange T (1999). "Mammalian telomeres end in a large duplex loop". Cell 97 (4): 503–14.
- ^ Burge S, Parkinson G, Hazel P, Todd A, Neidle S (2006). "Quadruplex DNA: sequence, topology and structure". Nucleic Acids Res 34 (19): 5402–15.
- ^ a b c d e Eisenberg DTA (2011). "An evolutionary review of human telomere biology: The thrifty telomere hypothesis and notes on potential adaptive paternal effects". American Journal of Human Biology. 23 (2): 149–167. doi:10.1002/ajhb.21127.
- ^ Sample, Ian (November 28, 2010). "Harvard scientists reverse the ageing process in mice – now for humans". The Guardian. London.
- ^ http://www.nature.com/nature/journal/vaop/ncurrent/full/nature09603.html
- ^ Joeng KS, Song EJ, Lee KJ, Lee J (2004). "Long lifespan in worms with long telomeric DNA". Nature Genetics. 36 (6): 607–11. doi:10.1038/ng1356. PMID 15122256.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nakagawa S, Gemmell NJ, Burke T (2004). "Measuring vertebrate telomeres: applications and limitations". Mol. Ecol. 13 (9): 2523–33. doi:10.1111/j.1365-294X.2004.02291.x. PMID 15315667.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Juola, Frans A; Haussmann, Mark F; Dearborn, Donald C; Vleck, Carol M (2006). "Telomere shortening in a long-lived marine bird: Cross-sectional analysis and test of an aging tool". The Auk. 123 (3): 775. doi:10.1642/0004-8038(2006)123[775:TSIALM]2.0.CO;2. ISSN 0004-8038.
- ^ Willeit Peter, Willeit Johann, Mayr Anita, Weger Siegfried, Oberhollenzer Friedrich, Brandstätter Anita, Kronenberg Florian, Kiechl Stefan (2010). "Telomere length and risk of incident cancer and cancer mortality". JAMA. 304 (1): 69–75. doi:10.1001/jama.2010.897. PMID 20606151.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Henson JD, Neumann AA, Yeager TR, Reddel RR (2002). "Alternative lengthening of telomeres in mammalian cells". Oncogene. 21 (4): 598–610. doi:10.1038/sj.onc.1205058. PMID 11850785.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chris Molenaar, Karien Wiesmeijer, Nico P. Verwoerd, Shadi Khazen, Roland Eils, Hans J. Tanke, and Roeland W. Dirks (2003-12-15). "Visualizing telomere dynamics in living mammalian cells using PNA probes". The EMBO Journal. 22 (24). The European Molecular Biology Organization: 6631–6641. doi:10.1093/emboj/cdg633. PMC 291828. PMID 14657034.
{{cite journal}}:|access-date=requires|url=(help)CS1 maint: multiple names: authors list (link) - ^ Philippi C, Loretz B, Schaefer UF, Lehr CM. (2010). "Telomerase as an emerging target to fight cancer - Opportunities and challenges for nanomedicine". Journal of Controlled Releases. 146 (2): 228–40. doi:10.1016/j.jconrel.2010.03.025. PMID 20381558.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Chin L, Artandi SE, Shen Q; et al. (1999). "p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis". Cell. 97 (4): 527–38. doi:10.1016/S0092-8674(00)80762-X. PMID 10338216.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Greenberg RA, Chin L, Femino A; et al. (1999). "Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse". Cell. 97 (4): 515–25. doi:10.1016/S0092-8674(00)80761-8. PMID 10338215.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Allshire RC et al. (1989) Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 17, 4611-4627.
- ^ Rufer N et al. (1998) Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 16: 743-747.
- ^ Cawthon RM. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res. 30(10):e47. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC115301/pdf/gnf046.pdf
Further reading
- Aubert G., Lansdorp P.M. (2008). "Telomeres and Aging". Physiological Reviews. 88 (2): 557–579. doi:10.1152/physrev.00026.2007. PMID 18391173.
{{cite journal}}: Unknown parameter|month=ignored (help)[1] - Cong YS, Wright WE, Shay JW (2002). "Human telomerase and its regulation". Microbiol. Mol. Biol. Rev. 66 (3): 407–25, table of contents. doi:10.1128/MMBR.66.3.407-425.2002. PMC 120798. PMID 12208997.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - Eisenberg DTA (2011). "An evolutionary review of human telomere biology: The thrifty telomere hypothesis and notes on potential adaptive paternal effects". American Journal of Human Biology: n/a–n/a. doi:10.1002/ajhb.21127.
- Tomaska, L., Nosek, J., Kramara, J., Griffith J.D. (2009). Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 16: 1010-1015.
- Weinstein BS, Ciszek D (2002). "The reserve-capacity hypothesis: evolutionary origins and modern implications of the trade-off between tumor-suppression and tissue-repair". Exp. Gerontol. 37 (5): 615–27. doi:10.1016/S0531-5565(02)00012-8. PMID 11909679.
{{cite journal}}: Unknown parameter|month=ignored (help) — A paper detailing the evolutionary origins and medical implications of the vertebrate telomere system, including the pervasive trade-off between cancer prevention and damage repair. Also addresses the probable danger posed by the elongation of telomeres in lab mice.
External links
- Telomeres and Telomerase - A summary
- Telomeres and Telomerase: Their Implications in Human Health and Disease online lecture by Elizabeth Blackburn
- Telomeres and Telomerase: The Means to the End Nobel Lecture by Elizabeth Blackburn, which includes a reference to the impact of stress, and pessimism on telomere length
- Telomerase and the Consequences of Telomere Dysfunction Nobel Lecture by Carol Greider
- DNA Ends: Just the Beginning Nobel Lecture by Jack Szostak
- Telome Health, Inc.: telomere science company founded by Elizabeth Blackburn
