Histone H2B
Histone H2B is one of the 4 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.[1]
Structure
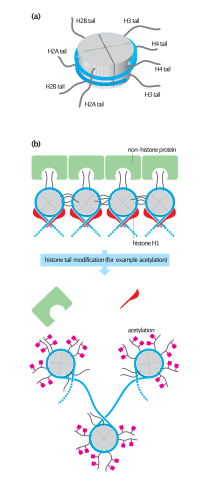
Histone H2B is a lightweight structural protein made of 126 amino acids.[2] Many of these amino acids have a positive charge at cellular pH, which allows them to interact with the negatively charged phosphate groups in DNA.[3] Along with a central globular domain, histone H2B has two[verification needed] flexible histone tails that extend outwards – one at the N-terminal end and one at C-terminal end. These are highly involved in condensing chromatin from the beads-on-a-string conformation to a 30-nm fiber.[3] Similar to other histone proteins, histone H2B has a distinct histone fold that is optimized for histone-histone as well as histone-DNA interactions.[1]
Two copies of histone H2B come together with two copies each of histone H2A, histone H3, and histone H4 to form the octamer core of the nucleosome[2] to give structure to DNA.[3] To facilitate this formation, histone H2B first binds to histone H2A to form a heterodimer.[2] Two of these heterodimers then bind together with a heterotetramer made of histone H3 and histone H4, giving the nucleosome its characteristic disk shape.[3] DNA is then wrapped around the entire nucleosome in groups of approximately 160 base pairs of DNA.[1] The wrapping continues until all chromatin has been packaged with the nucleosomes.[4]
Function

Histone H2B is a structural protein that helps organize eukaryotic DNA.[5] It plays an important role in the biology of the nucleus where it is involved in the packaging and maintaining of chromosomes,[5] regulation of transcription, and replication and repair of DNA.[2] Histone H2B helps regulate chromatin structure and function through post-translational modifications and specialized histone variants.[4]
Acetylation and ubiquitination are examples of two post-translational modifications that affect the function of histone H2B in particular ways. Hyperacetylation of histone tails helps DNA-binding proteins access chromatin by weakening histone-DNA and nucleosome-nucleosome interactions.[6] Furthermore, acetylation of a specific lysine residue binds to bromine-containing domains of certain transcription and chromatin regulatory proteins. This docking facilitates the recruitment of these proteins to the correct region of the chromosome. Ubiquitinated histone H2B is often found in regions of active transcription.[6] Through the facilitation of chromatin remodeling, it stimulates transcriptional elongation and sets the stage for further modifications that regulate multiple elements of transcription.[6] Specifically, the ubiquitin on histone H2B opens up and unfolds regions of chromatin allowing transcription machinery access to the promoter and coding regions of DNA.[7]
While only a few isoforms of histone H2B have been studied in depth, researchers have found that histone H2B variants serve important roles. If certain variants stopped functioning, centromeres would not form correctly, genome integrity would be lost, and the DNA damage response would be silenced.[4] Specifically, in some lower eukaryotes, a histone H2B variant binds to a histone H2A variant called H2AZ, localizes to active genes, and supports transcription in those regions. In mice, a variant called H2BE helps control the expression of olfactory genes. This supports the idea that isoforms of histone H2B may have specialized functions in different tissues.[4]
Isoforms
There are sixteen variants of histone H2B found in humans, thirteen of which are expressed in regular body cells and three of which are only expressed in the testes. These variants, also called isoforms, are proteins that are structurally very similar to the regular histone H2B but feature some specific variations in their amino acid sequence.[4] All variants of histone H2B contain the same number of amino acids, and the variations in sequence are few in number. Only two to five amino acids are changed, but even these small differences can alter the isoform’s, higher level structure.[4]
Histone H2B isoforms interact differently with other proteins, are found in specific regions of chromatin, have different types and numbers of post-translational modifications, and are more or less stable than regular histone H2B. All of these differences accumulate and cause the isoforms to have unique functions and even function differently in different tissues.[4]
Many histone H2B isoforms are expressed in a DNA replication independent manner. They are produced at the same level during all phases of the cell cycle. Regular histone H2B is only added to nucleosomes during the S-phase of the cell cycle when DNA is replicated; histone H2B isoforms can be added to nucleosomes at other times during the cell cycle.[4] Histone variants of H2B can be explored using "HistoneDB with Variants" database.
Post-Translational Modifications
Histone H2B is modified by a combination of several types of post-translational modifications.[1] These modifications affect the structural and functional organization of chromatin,[8] and the majority of them are found outside the globular domain of the nucleosome where amino acid residues are more accessible.[7] Possible modifications include acetylation, methylation, phosphorylation, ubiquitination, and sumoylation.[8] Acetylation, phosphorylation, and ubiquitination are the most common and most studied modifications of histone H2B.
Acetylation
Histone H2B proteins found both in the promoter and coding regions of genes contain specific patterns of hyperacetylation and hypoacetylation on certain lysine residues found in the N-terminal tail.[8] Acetylation relies on specific histone acetyltransferases that work at gene promoters during transcriptional activation.[1] Adding an acetyl group to lysine residues in one of several positions in the amino acid tail contributes to the activation of transcription.[3] In fact, scientists consider acetylation of histone H2B’s N-terminal tails, such as H2BK5ac, to be an extremely important part of regulating gene transcription.[8]
O-GlcNAcylation
Modification of H2B S112 with O-GlcNAc is thought to facilitate monoubiquitination of K112, which in turn is associated with transcriptionally activated regions.[9]
Phosphorylation
In histone H2B, a phosphorylated serine or threonine residue activates transcription.[3] When a cell experiences metabolic stress, an AMP-activated protein kinase phosphorylates the lysine at position 36 in histone H2B of the promoter and coding regions on DNA, which helps regulate transcriptional elongation.[2] If cells receive multiple apoptotic stimuli, caspase-3 activates the Mst1 kinase, which phosphorylates the serine at position 14 in all histone H2B proteins, which helps facilitate chromatin condensation. DNA damage can induce this same response on a more localized scale very quickly to help facilitate DNA repair.[2]
Ubiquitination
Ubiquitin residues are usually added to the lysine at position 120 on histone H2B. Ubiquitinating this lysine residue activates transcription.[3] Scientists have discovered other ubiquitination sites in recent years, but they are not well studied or understood at this time.[4] Ubiquitin-conjugating enzymes and ubiquitin ligases regulate the ubiquitination of histone H2B. These enzymes use co-transcription to conjugate ubiquitin to histone H2B. Histone H2B’s level of ubiquitination varies throughout the cell cycle. All ubiquitin moieties are removed from histone H2B during metaphase and re-conjugated during anaphase.[7]
Genetics
Histone H2B’s amino acid sequence is highly evolutionarily conserved. Even distantly related species have extremely similar histone H2B proteins.[3] The histone H2B family contains 214 members from many different and diverse species. In humans, histone H2B is coded for by twenty-three different genes,[10] none of which contain introns.[2] All of these genes are located in histone cluster 1 on chromosome 6 and cluster 2 and cluster 3 on chromosome 1. In each gene cluster, histone H2B genes share a promoter region with sequences that code for histone H2A. While all genes in the histone cluster are transcribed at high levels during S-phase, individual histone H2B genes are also expressed at other times during the cell cycle. They are dually regulated by the cluster promoter sequences and their specific promoter sequences.[4]
See also
- Histone code
- Nucleosome
- Chromatin
- Other histone proteins:
References
- ^ a b c d e Bhasin M, Reinherz EL, Reche PA (2006). "Recognition and classification of histones using support vector machine" (PDF). Journal of Computational Biology. 13 (1): 102–12. doi:10.1089/cmb.2006.13.102. PMID 16472024.
- ^ a b c d e f g Rønningen T, Shah A, Oldenburg AR, Vekterud K, Delbarre E, Moskaug JØ, Collas P (December 2015). "Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B". Genome Research. 25 (12): 1825–35. doi:10.1101/gr.193748.115. PMC 4665004. PMID 26359231.
- ^ a b c d e f g h Lodish HF, Berk A, Kaiser C, Krieger M, Bretscher A, Ploegh HL, Amon A, Scott MP (2013). Molecular Cell Biology (Seventh ed.). New York: Freeman. pp. 258–328. ISBN 978-1-4292-3413-9.
- ^ a b c d e f g h i j Molden RC, Bhanu NV, LeRoy G, Arnaudo AM, Garcia BA (2015). "Multi-faceted quantitative proteomics analysis of histone H2B isoforms and their modifications". Epigenetics & Chromatin. 8 (15): 15. doi:10.1186/s13072-015-0006-8. PMC 4411797. PMID 25922622.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Wu J, Mu S, Guo M, Chen T, Zhang Z, Li Z, Li Y, Kang X (January 2016). "Histone H2B gene cloning, with implication for its function during nuclear shaping in the Chinese mitten crab, Eriocheir sinensis". Gene. 575 (2 Pt 1): 276–84. doi:10.1016/j.gene.2015.09.005. PMID 26343795.
- ^ a b c "Histone H2B (53H3) Mouse mAb #2934". Cell Signaling Technology. Retrieved 24 November 2015.
- ^ a b c Osley MA (September 2006). "Regulation of histone H2A and H2B ubiquitylation". Briefings in Functional Genomics & Proteomics. 5 (3): 179–89. doi:10.1093/bfgp/ell022. PMID 16772277.
- ^ a b c d Golebiowski F, Kasprzak KS (November 2005). "Inhibition of core histones acetylation by carcinogenic nickel(II)". Molecular and Cellular Biochemistry. 279 (1–2): 133–9. doi:10.1007/s11010-005-8285-1. PMID 16283522.
- ^ R, Fujiki; W, Hashiba; H, Sekine; A, Yokoyama; T, Chikanishi; S, Ito; Y, Imai; J, Kim; Hh, He (2011-11-27). "GlcNAcylation of Histone H2B Facilitates Its Monoubiquitination". Nature. PMID 22121020. Retrieved 2020-06-01.
- ^ Khare SP, Habib F, Sharma R, Gadewal N, Gupta S, Galande S. "Histone H2B". HIstome: A Relational Knowledgebase of Human Proteins and Histone Modifying Enzymes. Nucleic Acids Research. Archived from the original on 16 October 2016. Retrieved 24 November 2015.
