Planctomycetota
| Planctomycetes | |
|---|---|
| |
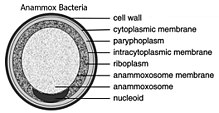
| cell diagram of Brocadia anammoxidans | |
| Scientific classification | |
| Domain: | |
| Superphylum: | |
| Phylum: | Planctomycetes Garrity & Holt 2001
|
| Classes | |
| Synonyms | |
| |
Planctomycetes are a phylum of aquatic bacteria and are found in samples of brackish, and marine and fresh water. They reproduce by budding. In structure, the organisms of this group are ovoid and have a holdfast, at the tip of a thin cylindrical extension from the cell body called the stalk, at the nonreproductive end that helps them to attach to each other during budding.
Cavalier-Smith has postulated that the Planctomycetes are within the clade Planctobacteria in the larger clade Gracilicutes, but this is not generally accepted.
Structure
For a long time bacteria belonging to this group were considered to lack peptidoglycan, (also called murein) in their cell walls, which is an important heteropolymer present in most bacterial cell walls that serves as a protective component. It was thought that instead their walls were made up of glycoprotein which is rich in glutamate. Recently, however, representatives of all three clades within the Planctomycetes were found to possess peptidoglycan-containing cell walls.[1][2]
Planctomycetes have a distinctive morphology with the appearance of membrane-bound internal compartments, often referred to as the paryphoplasm (ribosome-free space), pirellulosome (ribosome-containing space) and nucleoid (condensed nucleic acid region, in these species surrounded by a double membrane).[3][4] Until the discovery of the Poribacteria, planctomycetes were the only bacteria known with these apparent internal compartments.[5] Three-dimensional electron tomography reconstruction of a representative species, Gemmata obscuriglobus, has yielded varying interpretations of this observation. One 2013 study found the appearance of internal compartments to be due to a densely invaginated but continuous single membrane, concluding that only the two compartments typical of Gram-negative bacteria - the cytoplasm and periplasm - are present. However, the excess membrane triples the surface area of the cell relative to its volume, which may be related to Gemmata's sterol biosynthesis abilities.[6] A 2014 study using similar methods reported confirmation of the earlier enclosed compartment hypothesis.[7]
It has recently been shown that Gemmata obscuriglobus is able to take in large molecules via a process which in some ways resembles endocytosis, the process used by eukaryotic cells to engulf external items.[8][9]
Molecular Signatures
Although the Planctomycetes are renowned for their unusual cellular characteristics, their distinctness from all other bacteria is additionally supported by the shared presence of two conserved signature indels (CSIs).[10] These CSIs demarcate the group from neighboring phyla within the PVC group.[11] An additional CSI has been found that is shared by all Planctomycetes species, with the exception of Kuenenia stuttgartiensis, which is in line with the observation that K. stuttgartiensis forms a deep branch within the phylum.
A conserved signature indel has also been found to be shared by the entire PVC group, including Planctomycetes.[10][11] Planctomycetes also harbours an important conserved signature protein that has been characterized to play an important housekeeping function that is exclusive to members belonging to the PVC clade.[12]
Life cycle
The life cycle of many planctomycetes involves alternation between sessile cells and flagellated swarmer cells. The sessile cells bud to form the flagellated swarmer cells which swim for a while before settling down to attach and begin reproduction.
Phylogeny
The phylogeny based on the work of the All-Species Living Tree Project.[13]
| |||||||||||||
Taxonomy
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LSPN)[14] and the National Center for Biotechnology Information (NCBI).[15]
- Phylum "Planctomycetes" Garrity & Holt 2001 emend. Ward 2011 [Planctomycetaeota Oren et al. 2015]
- Class Phycisphaerae Fukunaga et al. 2010
- Order Tepidisphaerales Kovaleva et al. 2015
- Family Tepidisphaeraceae Kovaleva et al. 2015
- Genus Tepidisphaera Kovaleva et al. 2015
- Species Tepidisphaera mucosa Kovaleva et al. 2015
- Genus Tepidisphaera Kovaleva et al. 2015
- Family Tepidisphaeraceae Kovaleva et al. 2015
- Order Phycisphaerales Fukunaga et al. 2010
- Family Phycisphaeraceae Fukunaga et al. 2010
- Genus Algisphaera Yoon, Jang & Kasai 2014
- Species Algisphaera agarilytica Yoon, Jang & Kasai 2014
- Genus Phycisphaera Fukunaga et al. 2010
- Species Phycisphaera mikurensis Fukunaga et al. 2010
- Genus Algisphaera Yoon, Jang & Kasai 2014
- Family Phycisphaeraceae Fukunaga et al. 2010
- Order Tepidisphaerales Kovaleva et al. 2015
- Class Planctomycetacia Cavalier-Smith 2002
- Species ?"Candidatus Nostocoida limicola III" ♠ Liu et al. 2001
- Species ?"Candidatus Nostocoida acidiphila" ♠ Kulichevskaya et al. 2012
- Order "Brocadiales" Jetten et al. 2011
- Family "Brocadiaceae" Jetten et al. 2011
- Genus "Candidatus Anammoxoglobus" Kartal et al. 2006
- Species "Candidatus Anammoxoglobus propionicus" Kartal et al. 2006
- Genus "Candidatus Brocadia" Jetten et al. 2001
- Species "Candidatus B. anammoxidans" Jetten et al. 2001
- Species "Candidatus B. brasiliensis" Araujo et al. 2011
- Species "Candidatus B. caroliniensis"
- Species "Candidatus B. fulgida" Kartal et al. 2004
- Species "Candidatus B. sinica" Hu et al. 2010
- Genus "Candidatus Jettenia" Quan et al. 2008
- Species "Candidatus Jettenia asiatica" Quan et al. 2008
- Species "Candidatus Jettenia caeni" Ali et al. 2015
- Species "Candidatus Jettenia moscovienalis" Nikolaev et al. 2015
- Genus "Candidatus Kuenenia" Schmid et al. 2000
- Species "Candidatus Kuenenia stuttgartiensis" Schmid et al. 2000
- Genus "Candidatus Scalindua" Schmid et al. 2003
- Species "Candidatus S. arabica" Woebken et al. 2008
- Species "Candidatus S. marina" Van de Vossenberg et al. 2007
- Species "Candidatus S. profunda" Van De Vossenberg et al. 2008
- Species "Candidatus S. richardsii" Fuchsman et al. 2012
- Species "Candidatus S. wagneri" Schmid et al. 2003
- Species "Candidatus S. sorokinii" Kuypers et al. 2003
- Species Candidatus S. brodae Schmid et al. 2003
- Genus "Candidatus Anammoxoglobus" Kartal et al. 2006
- Family "Brocadiaceae" Jetten et al. 2011
- Order Planctomycetales Schlesner & Stackebrandt 1987 emend. Ward 2011
- Family Planctomycetaceae Schlesner & Stackebrandt 1987 emend. Scheuner et al. 2014 [Isosphaeraceae Kulichevskaya et al. 2015]
- Genus "Bythopirellula" ♠ Storesund & Ovreas 2013
- Species "Bythopirellula goksoyri" ♠ Storesund & Ovreas 2013
- Genus "Candidatus Anammoximicrobium" ♠ Khramenkov et al. 2013
- Species "Candidatus Anammoximicrobium moscowii" ♠ Khramenkov et al. 2013
- Genus Aquisphaera Bondoso et al. 2011
- Species Aquisphaera giovannonii Bondoso et al. 2011
- Genus Blastopirellula Schlesner et al. 2004
- Genus Gemmata Franzmann and Skerman 1985
- Species Gemmata obscuriglobus Franzmann & Skerman 1985
- Genus Gimesia Scheuner et al. 2015 [Planctomyces maris (ex Bauld & Staley 1976) Bauld & Staley 1980]
- Species Gimesia maris Scheuner et al. 2015 [Planctomyces maris (ex Bauld & Staley 1976) Bauld & Staley 1980]
- Genus Isosphaera Giovannoni et al. 1995
- Species Isosphaera pallida (ex Woronichin 1927) Giovannoni et al. 1995 nom. rev. ["Isocystis pallida" Woronichin 1927; "Torulopsidosira pallida" (Woronichin 1927) Geitler 1963]
- Genus Paludisphaera Kulichevskaya et al. 2015
- Species Paludisphaera borealis Kulichevskaya et al. 2015
- Genus Pirellula Schlesner & Hirsch 1987 emend. Schlesner et al. 2004 [Pirella Schlesner & Hirsch 1984 non Bainier 1883]
- Species Pirellula staleyi Schlesner & Hirsch 1987 [Pirella staleyi Schlesner & Hirsch 1984 ill.]
- Genus Planctomicrobium Kulichevskaya et al. 2015
- Species Planctomicrobium piriforme Kulichevskaya et al. 2015
- Genus Planctomyces Gimesi 1924
- Species P. bekefii ♪ Gimesi 1924 (type sp.) ["Blastocaulis sphaerica" Henrici & Johnson 1935; "Acinothrix globulifera" Novácek 1938; "Planctomyces subulatus" Wawrik 1952; "Planctomyces crassus" Hortobágyi 1965]
- Species P. guttaeformis ♪ (ex Hortobágyi 1965) Starr and Schmidt 1984
- Species P. stranskae ♪ (ex Wawrik 1952) Starr and Schmidt 1984
- Genus Planctopirus Scheuner et al. 2015
- Species Planctopirus limnophila corrig. (Hirsch & Muller 1986) Scheuner et al. 2015 [Planctomyces limnophilus Hirsch and Müller 1986]
- Genus Rhodopirellula Schlesner et al. 2004
- Species R. baltica Schlesner et al. 2004 (type sp.)
- Species R. caenicola Yoon et al. 2015
- Species R. rosea Roh et al. 2014
- Species R. rubra Bondoso et al. 2014
- Species R. lusitana Bondoso et al. 2014
- Species "R. europaea" ♠ Frank 2011
- Species "R. islandica" ♠ Kizina et al. 2015
- Species "R. maiorica" ♠ Frank 2011
- Species "R. sallentina" ♠ Frank 2011
- Genus Roseimaritima Bondoso et al. 2015
- Species Roseimaritima ulvae Bondoso et al. 2015
- Species Roseimaritima sediminicola kumar et al. 2020
- Genus Rubinisphaera Scheuner et al. 2015
- Species Rubinisphaera brasiliensis (Schlesner 1990) Scheuner et al. 2015 [Planctomyces brasiliensis Schlesner 1990]
- Genus Rubripirellula Bondoso et al. 2015
- Species Rubripirellula obstinata Bondoso et al. 2015
- Genus Schlesneria Kulichevskaya et al. 2007
- Species Schlesneria paludicola Kulichevskaya et al. 2007
- Genus Singulisphaera Kulichevskaya et al. 2008 emend. Kulichevskaya et al. 2012
- Species "S. mucilaginosa" ♠ Zaicnikova et al. 2011
- Species S. acidiphila Kulichevskaya et al. 2008 (type sp.)
- Species S. rosea Kulichevskaya et al. 2012
- Genus Telmatocola Kulichevskaya et al. 2012
- Species Telmatocola sphagniphila Kulichevskaya et al. 2012
- Genus Thermogutta Slobodkina et al. 2015
- Species T. hypogea Slobodkina et al. 2015
- Species T. terrifontis Slobodkina et al. 2015
- Genus Thermostilla Slobodkina et al. 2015
- Species Thermostilla marina Slobodkina et al. 2015
- Genus Zavarzinella Kulichevskaya et al. 2009
- Species Zavarzinella formosa Kulichevskaya et al. 2009
- Genus "Bythopirellula" ♠ Storesund & Ovreas 2013
- Family Planctomycetaceae Schlesner & Stackebrandt 1987 emend. Scheuner et al. 2014 [Isosphaeraceae Kulichevskaya et al. 2015]
- Class Phycisphaerae Fukunaga et al. 2010
Notes:
♠ Strains found at the National Center for Biotechnology Information (NCBI) but not listed in the List of Prokaryotic names with Standing in Nomenclature (LSPN)
♪ Prokaryotes where no pure (axenic) cultures are isolated or available, i. e. not cultivated or can not be sustained in culture for more than a few serial passages
See also
- Anammox, an anaerobic ammonium oxidation process, only found in the bacteria of this phylum
- List of bacterial orders
References
- ^ "Planctomycetes do possess a peptidoglycan cell wall". Nature Communications. 6: 7116. 2015. Bibcode:2015NatCo...6.7116J. doi:10.1038/ncomms8116. PMC 4432640. PMID 25964217.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ "Anammox Planctomycetes have a peptidoglycan cell wall". Nature Communications. 6: 6878. 2015. Bibcode:2015NatCo...6.6878V. doi:10.1038/ncomms7878. PMC 4432595. PMID 25962786.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Lindsay, M. R.; Webb, R. I.; Strous, M; Jetten, M. S.; Butler, M. K.; Forde, R. J.; Fuerst, J. A. (2001). "Cell compartmentalisation in planctomycetes: Novel types of structural organisation for the bacterial cell". Archives of Microbiology. 175 (6): 413–29. doi:10.1007/s002030100280. PMID 11491082.
- ^ Glöckner, F. O.; Kube, M; Bauer, M; Teeling, H; Lombardot, T; Ludwig, W; Gade, D; Beck, A; Borzym, K; Heitmann, K; Rabus, R; Schlesner, H; Amann, R; Reinhardt, R (2003). "Complete genome sequence of the marine planctomycete Pirellula sp. Strain 1". Proceedings of the National Academy of Sciences. 100 (14): 8298–303. Bibcode:2003PNAS..100.8298G. doi:10.1073/pnas.1431443100. PMC 166223. PMID 12835416.
- ^ Fieseler, L; Horn, M; Wagner, M; Hentschel, U (June 2004). "Discovery of the novel candidate phylum "Poribacteria" in marine sponges". Applied and Environmental Microbiology. 70 (6): 3724–32. doi:10.1128/aem.70.6.3724-3732.2004. PMC 427773. PMID 15184179.
- ^ "Three-Dimensional Reconstruction of Bacteria with a Complex Endomembrane System". PLOS Biology. 11 (5): e1001565. 2013. doi:10.1371/journal.pbio.1001565. PMC 3660258. PMID 23700385.
{{cite journal}}: Unknown parameter|authors=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Sagulenko, E; Morgan, G. P.; Webb, R. I.; Yee, B; Lee, K. C.; Fuerst, J. A. (2014). "Structural studies of planctomycete Gemmata obscuriglobus support cell compartmentalisation in a bacterium". PLOS ONE. 9 (3): e91344. Bibcode:2014PLoSO...991344S. doi:10.1371/journal.pone.0091344. PMC 3954628. PMID 24632833.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lonhienne, Thierry G. A.; Sagulenko, Evgeny; Webb, Richard I.; Lee, Kuo-Chang; Franke, Josef; Devos, Damien P.; Nouwens, Amanda; Carroll, Bernard J.; Fuerst, John A. (2010). "Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus". Proceedings of the National Academy of Sciences. 107 (29): 12883–12888. Bibcode:2010PNAS..10712883L. doi:10.1073/pnas.1001085107. PMC 2919973. PMID 20566852.
{{cite journal}}: Unknown parameter|lastauthoramp=ignored (|name-list-style=suggested) (help) - ^ Williams, Caroline (2011). "Who are you calling simple?". New Scientist. 211 (2821): 38–41. doi:10.1016/S0262-4079(11)61709-0.
- ^ a b Gupta RS, Bhandari V, Naushad HS (2012). "Molecular Signatures for the PVC Clade (Planctomycetes, Verrucomicrobia, Chlamydiae, and Lentisphaerae) of Bacteria Provide Insights into Their Evolutionary Relationships". Front Microbiol. 3: 327. doi:10.3389/fmicb.2012.00327. PMC 3444138. PMID 23060863.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Gupta RS (2016). "Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification". FEMS Microbiol Rev. 40 (4): 520–53. doi:10.1093/femsre/fuw011. PMID 27279642.
- ^ Lagkouvardos I, Jehl MA, Rattei T, Horn M (2014). "Signature protein of the PVC superphylum". Appl Environ Microbiol. 80 (2): 440–445. doi:10.1128/AEM.02655-13. PMC 3911108. PMID 24185849.
- ^ "16S rRNA-based LTP release 123 (full tree)" (PDF). Silva Comprehensive Ribosomal RNA Database. Retrieved 2016-03-20.
- ^ J.P. Euzéby. "Planctomycetes". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2016-03-20.
- ^ Sayers; et al. "Planctomycetes". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-03-20.
