A-DNA
A-DNA is one of the possible double helical structures which DNA can adopt. A-DNA is thought to be one of three biologically active double helical structures along with B-DNA and Z-DNA. It is a right-handed double helix fairly similar to the more common B-DNA form, but with a shorter, more compact helical structure whose base pairs are not perpendicular to the helix-axis as in B-DNA. It was discovered by Rosalind Franklin, who also named the A and B forms. She showed that DNA is driven into the A form when under dehydrating conditions. Such conditions are commonly used to form crystals, and many DNA crystal structures are in the A form.[1] The same helical conformation occurs in double-stranded RNAs, and in DNA-RNA hybrid double helices.
Structure
A-DNA is fairly similar to B-DNA given that it is a right-handed double helix with major and minor grooves. However, as shown in the comparison table below, there is a slight increase in the number of base pairs (bp) per turn (resulting in a smaller twist angle), and smaller rise per base pair (making A-DNA 20-25% shorter than B-DNA). The major groove of A-DNA is deep and narrow, while the minor groove is wide and shallow. A-DNA is broader and apparently more compressed along its axis than B-DNA.[2]
Comparison geometries of the most common DNA forms
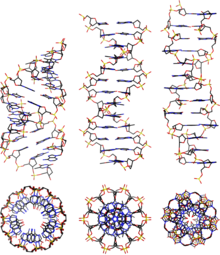

| Geometry attribute: | A-form | B-form | Z-form |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Repeating unit | 1 bp | 1 bp | 2 bp |
| Rotation/bp | 32.7° | 34.3° | 60°/2 |
| Mean bp/turn | 11 | 10 | 12 |
| Inclination of bp to axis | +19° | −1.2° | −9° |
| Rise/bp along axis | 2.6 Å (0.26 nm) | 3.4 Å (0.34 nm) | 3.7 Å (0.37 nm) |
| Rise/turn of helix | 28.6 Å (2.86 nm) | 35.7 Å (3.57 nm) | 45.6 Å (4.56 nm) |
| Mean propeller twist | +18° | +16° | 0° |
| Glycosyl angle | anti | anti | pyrimidine: anti, purine: syn |
| Nucleotide phosphate to phosphate distance | 5.9 Å | 7.0 Å | C: 5.7 Å, G: 6.1 Å |
| Sugar pucker | C3'-endo | C2'-endo | C: C2'-endo, G: C3'-endo |
| Diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
Biological function
Dehydration of DNA drives it into the A form, and this apparently protects DNA under conditions such as the extreme desiccation of bacteria.[3] Protein binding can also strip solvent off of DNA and convert it to the A form, as revealed by the structure of several hyperthermophilic archaeal viruses, including rod-shaped rudivirus SIRV2 [4], enveloped filamentous lipothrixviruses AFV1 [5] and SFV1 [6], tristromavirus PFV2 [7] as well as icosahedral portoglobovirus SPV1 [8]. A-form DNA is believed to be one of the adaptations of hyperthermophilic archaeal viruses to harsh environmental conditions in which these viruses thrive.
It has been proposed that the motors that package double-stranded DNA in bacteriophages exploit the fact that A-DNA is shorter than B-DNA, and that conformational changes in the DNA itself are the source of the large forces generated by these motors.[9] Experimental evidence for A-DNA as an intermediate in viral biomotor packing comes from double dye Förster resonance energy transfer measurements showing that B-DNA is shortened by 24% in a stalled ("crunched") A-form intermediate.[10][11] In this model, ATP hydrolysis is used to drive protein conformational changes that alternatively dehydrate and rehydrate the DNA, and the DNA shortening/lengthening cycle is coupled to a protein-DNA grip/release cycle to generate the forward motion that moves DNA into the capsid.
See also
References
- ^ Rosalind, Franklin (1953). "The Structure of Sodium Thymonucleate Fibres. I. The Influence of Water Content" (PDF). Acta Crystallographica. 6 (8): 673–677. doi:10.1107/s0365110x53001939.
- ^ Dickerson, Richard E. (1992). DNA Structure From A to Z. Vol. 211. pp. 67–111. doi:10.1016/0076-6879(92)11007-6. ISBN 9780121821128. PMID 1406328 – via Elsevier Science Direct.
{{cite book}}:|journal=ignored (help) - ^ Whelan DR, et al. (2014). "Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation". J R Soc Interface. 11 (97): 20140454. doi:10.1098/rsif.2014.0454. PMC 4208382. PMID 24898023.
- ^ Di Maio F, Egelman EH, et al. (2015). "A virus that infects a hyperthermophile encapsidates A-form DNA". Science. 348 (6237): 914–917. Bibcode:2015Sci...348..914D. doi:10.1126/science.aaa4181. PMC 5512286. PMID 25999507.
- ^ Kasson, P; DiMaio, F; Yu, X; Lucas-Staat, S; Krupovic, M; Schouten, S; Prangishvili, D; Egelman, EH (2017). "Model for a novel membrane envelope in a filamentous hyperthermophilic virus". eLife. 6: e26268. doi:10.7554/eLife.26268. PMC 5517147. PMID 28639939.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Liu, Y; Osinski, T; Wang, F; Krupovic, M; Schouten, S; Kasson, P; Prangishvili, D; Egelman, EH (2018). "Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile". Nature Communications. 9 (1): 3360. Bibcode:2018NatCo...9.3360L. doi:10.1038/s41467-018-05684-6. PMC 6105669. PMID 30135568.
- ^ Wang, F; Baquero, DP; Su, Z; Osinski, T; Prangishvili, D; Egelman, EH; Krupovic, M (2020). "Structure of a filamentous virus uncovers familial ties within the archaeal virosphere". Virus Evolution. 6 (1): veaa023. doi:10.1093/ve/veaa023. PMC 7189273. PMID 32368353.
- ^ Wang, F; Liu, Y; Su, Z; Osinski, T; de Oliveira, GAP; Conway, JF; Schouten, S; Krupovic, M; Prangishvili, D; Egelman, EH (2019). "A packing for A-form DNA in an icosahedral virus". Proceedings of the National Academy of Sciences of the United States of America. 116 (45): 22591–22597. doi:10.1073/pnas.1908242116. PMC 6842630. PMID 31636205.
- ^ Harvey, SC (2015). "The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages". Journal of Structural Biology. 189 (1): 1–8. doi:10.1016/j.jsb.2014.11.012. PMC 4357361. PMID 25486612.
- ^ Oram, M (2008). "Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure". J Mol Biol. 381 (1): 61–72. doi:10.1016/j.jmb.2008.05.074. PMC 2528301. PMID 18586272.
- ^ Ray, K (2010). "DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate". Virology. 398 (2): 224–232. doi:10.1016/j.virol.2009.11.047. PMC 2824061. PMID 20060554.
