Gene prediction

In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions. Gene finding is one of the first and most important steps in understanding the genome of a species once it has been sequenced.
In its earliest days, "gene finding" was based on painstaking experimentation on living cells and organisms. Statistical analysis of the rates of homologous recombination of several different genes could determine their order on a certain chromosome, and information from many such experiments could be combined to create a genetic map specifying the rough location of known genes relative to each other. Today, with comprehensive genome sequence and powerful computational resources at the disposal of the research community, gene finding has been redefined as a largely computational problem.
Determining that a sequence is functional should be distinguished from determining the function of the gene or its product. Predicting the function of a gene and confirming that the gene prediction is accurate still demands in vivo experimentation[1] through gene knockout and other assays, although frontiers of bioinformatics research [2] are making it increasingly possible to predict the function of a gene based on its sequence alone.
Gene prediction is one of the key steps in genome annotation, following sequence assembly, the filtering of non-coding regions and repeat masking.[3]
Gene prediction is closely related to the so-called 'target search problem' investigating how DNA-binding proteins (transcription factors) locate specific binding sites within the genome.[4][5] Many aspects of structural gene prediction are based on current understanding of underlying biochemical processes in the cell such as gene transcription, translation, protein–protein interactions and regulation processes, which are subject of active research in the various omics fields such as transcriptomics, proteomics, metabolomics, and more generally structural and functional genomics.
Empirical methods
[edit]In empirical (similarity, homology or evidence-based) gene finding systems, the target genome is searched for sequences that are similar to extrinsic evidence in the form of the known expressed sequence tags, messenger RNA (mRNA), protein products, and homologous or orthologous sequences. Given an mRNA sequence, it is trivial to derive a unique genomic DNA sequence from which it had to have been transcribed. Given a protein sequence, a family of possible coding DNA sequences can be derived by reverse translation of the genetic code. Once candidate DNA sequences have been determined, it is a relatively straightforward algorithmic problem to efficiently search a target genome for matches, complete or partial, and exact or inexact. Given a sequence, local alignment algorithms such as BLAST, FASTA and Smith-Waterman look for regions of similarity between the target sequence and possible candidate matches. Matches can be complete or partial, and exact or inexact. The success of this approach is limited by the contents and accuracy of the sequence database.
A high degree of similarity to a known messenger RNA or protein product is strong evidence that a region of a target genome is a protein-coding gene. However, to apply this approach systemically requires extensive sequencing of mRNA and protein products. Not only is this expensive, but in complex organisms, only a subset of all genes in the organism's genome are expressed at any given time, meaning that extrinsic evidence for many genes is not readily accessible in any single cell culture. Thus, to collect extrinsic evidence for most or all of the genes in a complex organism requires the study of many hundreds or thousands of cell types, which presents further difficulties. For example, some human genes may be expressed only during development as an embryo or fetus, which might be difficult to study for ethical reasons.
Despite these difficulties, extensive transcript and protein sequence databases have been generated for human as well as other important model organisms in biology, such as mice and yeast. For example, the RefSeq database contains transcript and protein sequence from many different species, and the Ensembl system comprehensively maps this evidence to human and several other genomes. It is, however, likely that these databases are both incomplete and contain small but significant amounts of erroneous data.
New high-throughput transcriptome sequencing technologies such as RNA-Seq and ChIP-sequencing open opportunities for incorporating additional extrinsic evidence into gene prediction and validation, and allow structurally rich and more accurate alternative to previous methods of measuring gene expression such as expressed sequence tag or DNA microarray.
Major challenges involved in gene prediction involve dealing with sequencing errors in raw DNA data, dependence on the quality of the sequence assembly, handling short reads, frameshift mutations, overlapping genes and incomplete genes.
In prokaryotes it's essential to consider horizontal gene transfer when searching for gene sequence homology. An additional important factor underused in current gene detection tools is existence of gene clusters — operons (which are functioning units of DNA containing a cluster of genes under the control of a single promoter) in both prokaryotes and eukaryotes. Most popular gene detectors treat each gene in isolation, independent of others, which is not biologically accurate.
Ab initio methods
[edit]Ab Initio gene prediction is an intrinsic method based on gene content and signal detection. Because of the inherent expense and difficulty in obtaining extrinsic evidence for many genes, it is also necessary to resort to ab initio gene finding, in which the genomic DNA sequence alone is systematically searched for certain tell-tale signs of protein-coding genes. These signs can be broadly categorized as either signals, specific sequences that indicate the presence of a gene nearby, or content, statistical properties of the protein-coding sequence itself. Ab initio gene finding might be more accurately characterized as gene prediction, since extrinsic evidence is generally required to conclusively establish that a putative gene is functional.
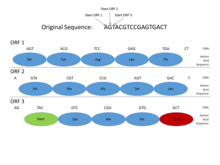
In the genomes of prokaryotes, genes have specific and relatively well-understood promoter sequences (signals), such as the Pribnow box and transcription factor binding sites, which are easy to systematically identify. Also, the sequence coding for a protein occurs as one contiguous open reading frame (ORF), which is typically many hundred or thousands of base pairs long. The statistics of stop codons are such that even finding an open reading frame of this length is a fairly informative sign. (Since 3 of the 64 possible codons in the genetic code are stop codons, one would expect a stop codon approximately every 20–25 codons, or 60–75 base pairs, in a random sequence.) Furthermore, protein-coding DNA has certain periodicities and other statistical properties that are easy to detect in a sequence of this length. These characteristics make prokaryotic gene finding relatively straightforward, and well-designed systems are able to achieve high levels of accuracy.
Ab initio gene finding in eukaryotes, especially complex organisms like humans, is considerably more challenging for several reasons. First, the promoter and other regulatory signals in these genomes are more complex and less well-understood than in prokaryotes, making them more difficult to reliably recognize. Two classic examples of signals identified by eukaryotic gene finders are CpG islands and binding sites for a poly(A) tail.
Second, splicing mechanisms employed by eukaryotic cells mean that a particular protein-coding sequence in the genome is divided into several parts (exons), separated by non-coding sequences (introns). (Splice sites are themselves another signal that eukaryotic gene finders are often designed to identify.) A typical protein-coding gene in humans might be divided into a dozen exons, each less than two hundred base pairs in length, and some as short as twenty to thirty. It is therefore much more difficult to detect periodicities and other known content properties of protein-coding DNA in eukaryotes.
Advanced gene finders for both prokaryotic and eukaryotic genomes typically use complex probabilistic models, such as hidden Markov models (HMMs) to combine information from a variety of different signal and content measurements. The GLIMMER system is a widely used and highly accurate gene finder for prokaryotes. GeneMark is another popular approach. Eukaryotic ab initio gene finders, by comparison, have achieved only limited success; notable examples are the GENSCAN and geneid programs. The GeneMark-ES and SNAP gene finders are GHMM-based like GENSCAN. They attempt to address problems related to using a gene finder on a genome sequence that it was not trained against.[7] [8] A few recent approaches like mSplicer,[9] CONTRAST,[10] or mGene[11] also use machine learning techniques like support vector machines for successful gene prediction. They build a discriminative model using hidden Markov support vector machines or conditional random fields to learn an accurate gene prediction scoring function.
Ab Initio methods have been benchmarked, with some approaching 100% sensitivity,[3] however as the sensitivity increases, accuracy suffers as a result of increased false positives.
Other signals
[edit]Among the derived signals used for prediction are statistics resulting from the sub-sequence statistics like k-mer statistics, Isochore (genetics) or Compositional domain GC composition/uniformity/entropy, sequence and frame length, Intron/Exon/Donor/Acceptor/Promoter and Ribosomal binding site vocabulary, Fractal dimension, Fourier transform of a pseudo-number-coded DNA, Z-curve parameters and certain run features.[12]
It has been suggested that signals other than those directly detectable in sequences may improve gene prediction. For example, the role of secondary structure in the identification of regulatory motifs has been reported.[13] In addition, it has been suggested that RNA secondary structure prediction helps splice site prediction.[14][15][16][17]
Neural networks
[edit]Artificial neural networks are computational models that excel at machine learning and pattern recognition. Neural networks must be trained with example data before being able to generalise for experimental data, and tested against benchmark data. Neural networks are able to come up with approximate solutions to problems that are hard to solve algorithmically, provided there is sufficient training data. When applied to gene prediction, neural networks can be used alongside other ab initio methods to predict or identify biological features such as splice sites.[18] One approach[19] involves using a sliding window, which traverses the sequence data in an overlapping manner. The output at each position is a score based on whether the network thinks the window contains a donor splice site or an acceptor splice site. Larger windows offer more accuracy but also require more computational power. A neural network is an example of a signal sensor as its goal is to identify a functional site in the genome.
Combined approaches
[edit]Programs such as Maker combine extrinsic and ab initio approaches by mapping protein and EST data to the genome to validate ab initio predictions. Augustus, which may be used as part of the Maker pipeline, can also incorporate hints in the form of EST alignments or protein profiles to increase the accuracy of the gene prediction.
Comparative genomics approaches
[edit]As the entire genomes of many different species are sequenced, a promising direction in current research on gene finding is a comparative genomics approach.
This is based on the principle that the forces of natural selection cause genes and other functional elements to undergo mutation at a slower rate than the rest of the genome, since mutations in functional elements are more likely to negatively impact the organism than mutations elsewhere. Genes can thus be detected by comparing the genomes of related species to detect this evolutionary pressure for conservation. This approach was first applied to the mouse and human genomes, using programs such as SLAM, SGP and TWINSCAN/N-SCAN and CONTRAST.[20]
Multiple informants
[edit]TWINSCAN examined only human-mouse synteny to look for orthologous genes. Programs such as N-SCAN and CONTRAST allowed the incorporation of alignments from multiple organisms, or in the case of N-SCAN, a single alternate organism from the target. The use of multiple informants can lead to significant improvements in accuracy.[20]
CONTRAST is composed of two elements. The first is a smaller classifier, identifying donor splice sites and acceptor splice sites as well as start and stop codons. The second element involves constructing a full model using machine learning. Breaking the problem into two means that smaller targeted data sets can be used to train the classifiers, and that classifier can operate independently and be trained with smaller windows. The full model can use the independent classifier, and not have to waste computational time or model complexity re-classifying intron-exon boundaries. The paper in which CONTRAST is introduced proposes that their method (and those of TWINSCAN, etc.) be classified as de novo gene assembly, using alternate genomes, and identifying it as distinct from ab initio, which uses a target 'informant' genomes.[20]
Comparative gene finding can also be used to project high quality annotations from one genome to another. Notable examples include Projector, GeneWise, GeneMapper and GeMoMa. Such techniques now play a central role in the annotation of all genomes.
Pseudogene prediction
[edit]Pseudogenes are close relatives of genes, sharing very high sequence homology, but being unable to code for the same protein product. Whilst once relegated as byproducts of gene sequencing, increasingly, as regulatory roles are being uncovered, they are becoming predictive targets in their own right.[21] Pseudogene prediction utilises existing sequence similarity and ab initio methods, whilst adding additional filtering and methods of identifying pseudogene characteristics.
Sequence similarity methods can be customised for pseudogene prediction using additional filtering to find candidate pseudogenes. This could use disablement detection, which looks for nonsense or frameshift mutations that would truncate or collapse an otherwise functional coding sequence.[22] Additionally, translating DNA into proteins sequences can be more effective than just straight DNA homology.[21]
Content sensors can be filtered according to the differences in statistical properties between pseudogenes and genes, such as a reduced count of CpG islands in pseudogenes, or the differences in G-C content between pseudogenes and their neighbours. Signal sensors also can be honed to pseudogenes, looking for the absence of introns or polyadenine tails. [23]
Metagenomic gene prediction
[edit]Metagenomics is the study of genetic material recovered from the environment, resulting in sequence information from a pool of organisms. Predicting genes is useful for comparative metagenomics.
Metagenomics tools also fall into the basic categories of using either sequence similarity approaches (MEGAN4) and ab initio techniques (GLIMMER-MG).
Glimmer-MG[24] is an extension to GLIMMER that relies mostly on an ab initio approach for gene finding and by using training sets from related organisms. The prediction strategy is augmented by classification and clustering gene data sets prior to applying ab initio gene prediction methods. The data is clustered by species. This classification method leverages techniques from metagenomic phylogenetic classification. An example of software for this purpose is, Phymm, which uses interpolated markov models—and PhymmBL, which integrates BLAST into the classification routines.
MEGAN4[25] uses a sequence similarity approach, using local alignment against databases of known sequences, but also attempts to classify using additional information on functional roles, biological pathways and enzymes. As in single organism gene prediction, sequence similarity approaches are limited by the size of the database.
FragGeneScan and MetaGeneAnnotator are popular gene prediction programs based on Hidden Markov model. These predictors account for sequencing errors, partial genes and work for short reads.
Another fast and accurate tool for gene prediction in metagenomes is MetaGeneMark.[26] This tool is used by the DOE Joint Genome Institute to annotate IMG/M, the largest metagenome collection to date.
See also
[edit]- List of gene prediction software
- Phylogenetic footprinting
- Protein function prediction
- Protein structure prediction
- Protein–protein interaction prediction
- Pseudogene (database)
- Sequence mining
- Sequence similarity (homology)
References
[edit]- ^ Sleator RD (August 2010). "An overview of the current status of eukaryote gene prediction strategies". Gene. 461 (1–2): 1–4. doi:10.1016/j.gene.2010.04.008. PMID 20430068.
- ^ Ejigu, Girum Fitihamlak; Jung, Jaehee (2020-09-18). "Review on the Computational Genome Annotation of Sequences Obtained by Next-Generation Sequencing". Biology. 9 (9): 295. doi:10.3390/biology9090295. ISSN 2079-7737. PMC 7565776. PMID 32962098.
- ^ a b Yandell M, Ence D (April 2012). "A beginner's guide to eukaryotic genome annotation". Nature Reviews. Genetics. 13 (5): 329–42. doi:10.1038/nrg3174. PMID 22510764. S2CID 3352427.
- ^ Redding S, Greene EC (May 2013). "How do proteins locate specific targets in DNA?". Chemical Physics Letters. 570: 1–11. Bibcode:2013CPL...570....1R. doi:10.1016/j.cplett.2013.03.035. PMC 3810971. PMID 24187380.
- ^ Sokolov IM, Metzler R, Pant K, Williams MC (August 2005). "Target search of N sliding proteins on a DNA". Biophysical Journal. 89 (2): 895–902. Bibcode:2005BpJ....89..895S. doi:10.1529/biophysj.104.057612. PMC 1366639. PMID 15908574.
- ^ Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl D (2015). Brock Biology of Microorganisms (14th ed.). Boston: Pearson. ISBN 9780321897398.
- ^ "GeneMark-ES".
- ^ Korf I (May 2004). "Gene finding in novel genomes". BMC Bioinformatics. 5: 59. doi:10.1186/1471-2105-5-59. PMC 421630. PMID 15144565.
- ^ Rätsch G, Sonnenburg S, Srinivasan J, Witte H, Müller KR, Sommer RJ, Schölkopf B (February 2007). "Improving the Caenorhabditis elegans genome annotation using machine learning". PLOS Computational Biology. 3 (2): e20. Bibcode:2007PLSCB...3...20R. doi:10.1371/journal.pcbi.0030020. PMC 1808025. PMID 17319737.
- ^ Gross SS, Do CB, Sirota M, Batzoglou S (2007-12-20). "CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction". Genome Biology. 8 (12): R269. doi:10.1186/gb-2007-8-12-r269. PMC 2246271. PMID 18096039.
- ^ Schweikert G, Behr J, Zien A, Zeller G, Ong CS, Sonnenburg S, Rätsch G (July 2009). "mGene.web: a web service for accurate computational gene finding". Nucleic Acids Research. 37 (Web Server issue): W312–6. doi:10.1093/nar/gkp479. PMC 2703990. PMID 19494180.
- ^ Saeys Y, Rouzé P, Van de Peer Y (February 2007). "In search of the small ones: improved prediction of short exons in vertebrates, plants, fungi and protists". Bioinformatics. 23 (4): 414–20. doi:10.1093/bioinformatics/btl639. PMID 17204465.
- ^ Hiller M, Pudimat R, Busch A, Backofen R (2006). "Using RNA secondary structures to guide sequence motif finding towards single-stranded regions". Nucleic Acids Research. 34 (17): e117. doi:10.1093/nar/gkl544. PMC 1903381. PMID 16987907.
- ^ Patterson DJ, Yasuhara K, Ruzzo WL (2002). "Pre-mRNA secondary structure prediction aids splice site prediction". Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing: 223–34. PMID 11928478.
- ^ Marashi SA, Goodarzi H, Sadeghi M, Eslahchi C, Pezeshk H (February 2006). "Importance of RNA secondary structure information for yeast donor and acceptor splice site predictions by neural networks". Computational Biology and Chemistry. 30 (1): 50–7. doi:10.1016/j.compbiolchem.2005.10.009. PMID 16386465.
- ^ Marashi SA, Eslahchi C, Pezeshk H, Sadeghi M (June 2006). "Impact of RNA structure on the prediction of donor and acceptor splice sites". BMC Bioinformatics. 7: 297. doi:10.1186/1471-2105-7-297. PMC 1526458. PMID 16772025.
- ^ Rogic, S (2006). The role of pre-mRNA secondary structure in gene splicing in Saccharomyces cerevisiae (PDF) (PhD thesis). University of British Columbia. Archived from the original (PDF) on 2009-05-30. Retrieved 2007-04-01.
- ^ Goel N, Singh S, Aseri TC (July 2013). "A comparative analysis of soft computing techniques for gene prediction". Analytical Biochemistry. 438 (1): 14–21. doi:10.1016/j.ab.2013.03.015. PMID 23529114.
- ^ Johansen, ∅Ystein; Ryen, Tom; Eftes∅l, Trygve; Kjosmoen, Thomas; Ruoff, Peter (2009). "Splice Site Prediction Using Artificial Neural Networks". Computational Intelligence Methods for Bioinformatics and Biostatistics. Lec Not Comp Sci. Vol. 5488. pp. 102–113. doi:10.1007/978-3-642-02504-4_9. ISBN 978-3-642-02503-7.
- ^ a b c Gross SS, Do CB, Sirota M, Batzoglou S (2007). "CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction". Genome Biology. 8 (12): R269. doi:10.1186/gb-2007-8-12-r269. PMC 2246271. PMID 18096039.
- ^ a b Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB (August 2010). "Annotating non-coding regions of the genome". Nature Reviews. Genetics. 11 (8): 559–71. doi:10.1038/nrg2814. PMID 20628352. S2CID 6617359.
- ^ Svensson O, Arvestad L, Lagergren J (May 2006). "Genome-wide survey for biologically functional pseudogenes". PLOS Computational Biology. 2 (5): e46. Bibcode:2006PLSCB...2...46S. doi:10.1371/journal.pcbi.0020046. PMC 1456316. PMID 16680195.
- ^ Zhang Z, Gerstein M (August 2004). "Large-scale analysis of pseudogenes in the human genome". Current Opinion in Genetics & Development. 14 (4): 328–35. doi:10.1016/j.gde.2004.06.003. PMID 15261647.
- ^ Kelley DR, Liu B, Delcher AL, Pop M, Salzberg SL (January 2012). "Gene prediction with Glimmer for metagenomic sequences augmented by classification and clustering". Nucleic Acids Research. 40 (1): e9. doi:10.1093/nar/gkr1067. PMC 3245904. PMID 22102569.
- ^ Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (September 2011). "Integrative analysis of environmental sequences using MEGAN4". Genome Research. 21 (9): 1552–60. doi:10.1101/gr.120618.111. PMC 3166839. PMID 21690186.
- ^ Zhu W, Lomsadze A, Borodovsky M (July 2010). "Ab initio gene identification in metagenomic sequences". Nucleic Acids Research. 38 (12): e132. doi:10.1093/nar/gkq275. PMC 2896542. PMID 20403810.
External links
[edit]- Augustus
- FGENESH Archived 2013-01-04 at archive.today
- GeMoMa - Homology-based gene prediction based on amino acid and intron position conservation as well as RNA-Seq data
- geneid, SGP2
- Glimmer Archived 2011-08-26 at the Wayback Machine, GlimmerHMM Archived 2011-08-18 at the Wayback Machine
- GenomeThreader
- ChemGenome
- GeneMark
- Gismo
- mGene
- StarORF — A multi-platform and web tool for predicting ORFs and obtaining reverse complement sequence
- Maker - A portable and easily configurable genome annotation pipeline
