Kaposi's sarcoma-associated herpesvirus
| Kaposi's sarcoma-associated herpesvirus | |
|---|---|
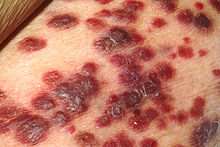
| |
| Kaposi's sarcoma | |
| Virus classification | |
| Group: | Group I (dsDNA)
|
| Order: | |
| Family: | |
| Subfamily: | |
| Genus: | |
| Species: | Human herpesvirus 8
|
Kaposi's sarcoma-associated herpesvirus (KSHV) is the eighth human herpesvirus; its formal name according to the International Committee on Taxonomy of Viruses (ICTV) is HHV-8. Like other herpesviruses, its informal name (KSHV) is used interchangeably with its ICTV name. This virus causes Kaposi's sarcoma, a cancer commonly occurring in AIDS patients,[1] as well as primary effusion lymphoma,[2] some types of multicentric Castleman's disease and KSHV inflammatory cytokine syndrome.[3] It is one of seven currently known human cancer viruses, or oncoviruses.[1]
History
In 1872, Moritz Kaposi described a blood vessel tumor[4] (originally called "idiopathic multiple pigmented sarcoma of the skin") that has since been eponymously named Kaposi's sarcoma (KS). KS was thought to be an uncommon tumor of Jewish and Mediterranean populations until it was realized that it is actually quite common throughout sub-Saharan Africa. This led to the first suggestions in the 1950s that this tumor might be caused by a virus. With the onset of the AIDS epidemic in the early 1980s, there was a sudden epidemic resurgence of KS affecting primarily gay and bisexual AIDS patients with up to 50% of reported AIDS patients having this tumor—an extraordinary rate of cancer predisposition. Careful analysis of epidemiologic data by Valerie Beral, Thomas Peterman and Harold Jaffe,[5] led these investigators to propose that KS is caused by an unknown sexually transmitted virus that rarely causes tumors unless the host becomes immunosuppressed, as in AIDS.

It was first found by Yuan Chang and Patrick S. Moore, a wife and husband team at Columbia University in 1994,[6][7][8] by isolating DNA fragments of a herpesvirus from a Kaposi's sarcoma (KS) tumor in an AIDS patient. As early as 1984, scientists had reported seeing herpesvirus-like structures in KS tumors by electron microscopy. Scientists by then had been searching for the agent causing Kaposi's sarcoma and over 20 agents had been described as the possible cause of KS, including cytomegalovirus and HIV itself. Chang and Moore used representational difference analysis, or RDA, to find KSHV by comparing KS tumor tissue from an AIDS patient to his own unaffected tissue. The idea behind this experiment was that if a virus causes KS, the genomic DNA in the two samples should be precisely identical except for DNA belonging to the virus. In their initial RDA experiment, they isolated two small DNA fragments that represented less than 1% of the actual viral genome. These fragments were similar to but distinct from known herpevirus sequences indicating the presence of a new virus. Starting from these fragments, this research team was then able to sequence the entire genome of the virus less than two years later.
The discovery of this herpesvirus sparked considerable controversy and scientific in-fighting until sufficient data had been collected to show that indeed KSHV was the causative agent of Kaposi's sarcoma.[9] The virus is now known to be a widespread infection of people living in sub-Saharan Africa; intermediate levels of infection occur in Mediterranean populations (including Israel, Saudi Arabia, Italy and Greece) and low levels of infection occur in most Northern European and North American populations. Gay and bi-sexual men are more susceptible to infection (through still unknown routes of sexual transmission) whereas the virus is transmitted through non-sexual routes in developing countries.[citation needed]
Virology
KSHV is a herpesvirus, and is a large double-stranded DNA virus with a protein covering that packages its nucleic acids, called the capsid, which is then surrounded by an amorphous protein layer called the tegument, and finally enclosed in a lipid envelope derived in part from the cell membrane. KSHV has a genome which is approximately 165,000 nucleic acid bases in length. It is a rhadinovirus, and is remarkable since it has stolen numerous genes from host cells including genes that encode for complement-binding protein, IL-6, BCL-2, cyclin-D, a G protein-coupled receptor, interferon regulatory factor and Flice inhibitory protein (FLIP), as well as DNA synthesis proteins including dihydrofolate reductase, thymidine kinase, thymidylate synthetase, DNA polymerase and many others. While no other human tumor virus possesses these same genes, other tumor viruses target the same cellular pathways illustrating that at a basic level, all tumor viruses appear to attack the same cellular control pathways, so-called tumor suppressor pathways.[citation needed]
After infection, the virus enters into lymphocytes via macropinosomes where it remains in a latent ("quiet") state expressing the viral latency-associated nuclear antigen, LANA. The virus exists as a naked circular piece of DNA called an episome and uses the cellular replication machinery to replicate itself. LANA tethers the viral DNA to cellular chromosomes, inhibits p53 and retinoblastoma protein and suppresses viral genes needed for full virus production and assembly ("lytic replication"). Various signals such as inflammation may provoke the virus to enter into lytic replication. When this occurs, the viral episome starts replicating itself in the form of linear DNA molecules that are packaged into virus particles which are expelled from the cell, to infect new cells or to be transmitted to a new host. When the virus enters into lytic replication, thousands of virus particles can be made from a single cell, which usually results in death of the infected cell.
The viral genome consists of a ~145 kbase long unique region, encoding all of expressed viral genes, which is flanked by ~20-30 kbases of terminal repeat sequences.[10] Each terminal repeat unit is 801 bp in length, has 85% G+C content and is oriented in a repetitive head-to-tail fashion. During latency, the virus genome depends on the host replication machinery and replicates as closed circular episome ("plasmid") using sequences within the terminal repeats as a replication origin. When the virus reactivates into lytic replication, it is believed that the virus genome is replicated as a continuous linear molecule off from an episome (so called rolling circle model). As each unit genome is replicated, it is cut within the terminal repeat region, and then packaged into a virus particle (virion). The virus then becomes enveloped with a lipid membrane as it transits the nucleus and the cytoplasm to exit the cell. Thus, whereas KSHV genome is circular in the nucleus of latently infected cells, it is packaged into infectious viruses as a linear molecule. Once the virus newly infects a cell, the lipid membrane is shed and the virion travels to the nucleus. The viral genome is released where it recircularizes through a poorly understood process that appears to involve homologous recombination.
The primary viral protein responsible for the switch between latent and lytic replication is the ORF50 Replication Transactivation Activator (RTA). When cell signaling conditions activate the generation of RTA, it in turn activates synthesis of a stereotypic cascade of secondary and tertiary viral proteins that ultimately make components of the virus capsid and also the DNA synthesis enzymes required to replicate the virus genome.[11]
Pathophysiology
The mechanisms by which the virus is contracted are not well understood. Healthy individuals can be infected with the virus and show no signs or symptoms, due to the immune system's ability to keep the infection in check. Infection is of particular concern to the immunocompromized. Cancer patients receiving chemotherapy, AIDS patients and organ transplant patients are all at a high risk of showing signs of infection.
Infection with this virus is thought to be lifelong, but a healthy immune system will keep the virus in check. Many people infected with KSHV will never show any symptoms. Kaposi's sarcoma occurs when someone who has been infected with KSHV becomes immunocompromised due to AIDS, medical treatment or very rarely aging.
KSHV is a known causative agent of four diseases:[3]
- Kaposi's sarcoma – an angioproliferative tumor that can involve skin (most often), lymph nodes, or viscera,
- KSHV associated multicentric Castleman's disease (MCD) – a B-cell lymphoproliferative disorder,
- Primary effusion lymphoma – KSHV-associated aggressive mature monoclonal B-cell lymphoma,
- KSHV inflammatory cytokine syndrome – a syndrome with MCD-like symptoms but without associated pathology.
Epidemiology
The seroprevalence of HHV-8 varies significantly geographically and infection rates in northern European, southeast Asian, and Caribbean countries are between 2-4%,[12] in Mediterranean countries at approximately 10%, and in sub-Saharan African countries at approximately 40%.[13] In South America, infection rates are low in general but are high among Amerindians.[14] Even within individual countries, significant variation can be observed across different regions, with infection rates of about 19.2% in Xinjiang compared to about 9.5% in Hubei, China.[15] Although seroprevalence has been consistently shown to increase with age in a linear manner,[15][16][17][18] countries with high infection rates may see higher seroprevalence in younger age groups.[19] Educational level has shown an inverse correlation with infection rates.[16][17] Individuals infected with HIV-1 or genital warts are generally more likely to be co-infected with HHV-8.[13][15][20][21]
In countries with low seroprevalence, HHV-8 is primarily limited to AIDS and KS patients.[22] In countries with high seroprevalence, infection is frequent in childhood,[23] indicating a likely mother-to-child transmission.[24] In a Zambian survey, all children with KS had mothers who were positive for HHV-8, whereas not all children whose mothers had KS were HHV-8 positive.[25] In another Zambian survey, 13.8% of children were seropositive for HHV-8 by age 4.[21] Seroprevalence has not been shown to vary significantly because of gender or marital status.[18]
Evolution
The most recent common ancestor of this virus has been estimated to have evolved 29,872 years (95% highest probability density 26,851-32,760 years) ago.[26] the most recent common ancestor for viruses isolated in Xinjiang was 2037 years (95% highest probability density 1843–2229 years) ago. Given the historical links between the Mediterranean and Xinjiang during the Roman period it seems likely that this virus was introduced to Xinjiang along the Silk Road. The mutation rate was estimated to be 3.44 × 10−6 substitutions per site per year (95% highest probability density 2.26 × 10-6 to 4.71 × 10−6).
Prevention
Since persons infected with KSHV will asymptomatically give the virus, caution should be used by sex partners in having unprotected sex and activities where saliva might be shared during sexual activity. Prudent advice is to use condoms when needed and avoid deep kissing with partners with KSHV and HIV infections or whose status is unknown. Blood tests to detect antibodies to virus exist.
Treatment
Kaposi's sarcoma is usually a localized tumor that can be treated either surgically or through local irradiation. Chemotherapy with drugs such as liposomal anthracyclines or paclitaxel may be used, particularly for invasive disease. Antiviral drugs, such as ganciclovir, that target the replication of herpesviruses such as KSHV have been used to successfully prevent development of Kaposi's sarcoma,[27] although once the tumor develops these drugs are of little or no use. For patients with AIDS-KS, the most effective therapy is highly active antiretroviral therapy to reduce HIV infection.[28] AIDS patients receiving adequate anti-HIV treatment may have up to a 90% reduction in Kaposi's sarcoma occurrence.
KSHV genes
KSHV encodes for ~90 genes and multiple non-coding RNAs. The "ORF" genes are named based on genome position of the homologous genes in the first rhadinovirus described, herpesvirus saimiri. The "K" genes are unique to KSHV, Some KSHV genes have well-characterized functions, while others remain uncharacterized.
ORF2 – dihydrofolate reductase
ORF8 – gB – envelope glycoprotein involved in viral entry
ORF9 – Pol8 – DNA polymerase required for viral DNA replication
ORF10 – regulates RNA export and responses to type I IFNs
ORF16 – vBcl2
ORF18, ORF24, ORF30, ORF31, ORF34, ORF66 – viral transcription factors required for the expression of late genes
ORF21 – vTK – thymidine kinase
ORF22 – gH – envelope glycoprotein involved in viral entry
ORF23 – uncharacterized
ORF25, ORF26 and ORF65 – capsid proteins
ORF33 – involved in viral particle formation
ORF34 – unclear function
ORF35 – unclear function, mutant does not express early viral genes
ORF36 – vPK – viral protein kinase with multiple roles in replication cycle
ORF37 – SOX – dual function protein – DNase activity required for genome packaging and RNase activity regulates host gene expression
ORF38 – involved in viral particle formation
ORF39 – gM – envelope glycoprotein
ORF40 and ORF41 – helicase and primase – DNA replication
ORF42 – uncharacterized
ORF45
ORF47 – gL – envelope glycoprotein involved in viral entry
ORF49 – may be required for viral gene expression
ORF50 – RTA – transcription factor driving lytic reactivation
ORF52 – KicGAS – tegument protein required for formation of virions and inhibition of cGAS
ORF53 – gN – envelope glycoprotein
ORF55 – uncharacterized
ORF57 – MTA – regulates RNA stability, export and translation of viral genes
ORF59 – PF–8 – polymerase processivity factor, accessory subunit of viral DNA polymerase
ORF67 and ORF69 – nuclear egress
ORF70 – thymidylate synthase
ORF72 – vCyclin
ORF73 – LANA – tethers genome to chromosome during latency, also regulates host gene expression
ORF74 – vGPCR
ORF75 – FGARAT
PAN – non–coding RNA
miRNAs (mirKs) – expressed during latency to regulate proliferation and cell death
K1 – involved in oncogenesis
K3 and K5 – ubiquitin E3 ligases – regulate antigen presentation
K4 – vCCL2 – chemokine
K4.1 – vCCL3 – chemokine
K8 – transcriptional repressor – modulates chromatin
K8.1 – envelope glycoprotein
K9 – vIRF1
K10 – vIRF4
K10.5 – vIRF3/LANA2 – expressed during latency in B cells
K11 – vIRF2
K12 – kaposin
K13 – vFLIP
See also
- Oncovirus (cancer virus)
References
- ^ a b Boshoff, C.; Weiss, R. (2002). "Aids-related malignancies". Nature Reviews Cancer. 2 (5): 373–382. doi:10.1038/nrc797. PMID 12044013.
- ^ Cesarman, E.; Chang, Y.; Moore, P. S.; Said, J. W.; Knowles, D. M. (1995). "Kaposi's Sarcoma–Associated Herpesvirus-Like DNA Sequences in AIDS-Related Body-Cavity–Based Lymphomas". New England Journal of Medicine. 332 (18): 1186–1191. doi:10.1056/NEJM199505043321802. PMID 7700311.
- ^ a b Goncalves, Priscila H.; Ziegelbauer, Joseph; Uldrick, Thomas S.; Yarchoan, Robert (2017). "Kaposi sarcoma herpesvirus-associated cancers and related diseases". Curr Opin HIV AIDS. 12: 47–56. doi:10.1097/COH.0000000000000330.
- ^ Kaposi, M (1872). "Idiopathisches multiples Pigmentsarkom der Haut". Archiv für Dermatologie und Syphilis. 4 (2): 265–273. doi:10.1007/BF01830024.
Translated in Kaposi, M (2008). "Idiopathic multiple pigmented sarcoma of the skin". CA: A Cancer Journal for Clinicians. 32 (6): 342. doi:10.3322/canjclin.32.6.342. - ^ Beral V, Peterman TA, Berkelman RL, Jaffe HW (1990). "Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection?". Lancet. 335 (8682): 123–8. doi:10.1016/0140-6736(90)90001-L. PMID 1967430.
- ^ Chang, Y.; Cesarman, E.; Pessin, M. S.; Lee, F.; Culpepper, J.; Knowles, D. M.; Moore, P. S. (1994). "Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma". Science. 266 (5192): 1865–1869. doi:10.1126/science.7997879. PMID 7997879.
- ^ Moore PS, Chang Y (May 1995). "Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection". The New England Journal of Medicine. 332 (18): 1181–5. doi:10.1056/NEJM199505043321801. PMID 7700310.
- ^ Antman, K.; Chang, Y. (2000). "Kaposi's Sarcoma". New England Journal of Medicine. 342 (14): 1027–1038. doi:10.1056/NEJM200004063421407. PMID 10749966.
- ^ Boshoff C, Weiss RA (May 2000). "Addressing Controversies Over Kaposi's Sarcoma". Journal of the National Cancer Institute. 92 (9): 677–679. doi:10.1093/jnci/92.9.677. PMID 10793096.
- ^ Russo JJ, Bohenzky RA, Chien MC, et al. (December 1996). "Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8)". Proceedings of the National Academy of Sciences of the United States of America. 93 (25): 14862–7. doi:10.1073/pnas.93.25.14862. PMC 26227. PMID 8962146.
- ^ Bu W, Palmeri D, Krishnan R, et al. (November 2008). "Identification of Direct Transcriptional Targets of the Kaposi's Sarcoma-Associated Herpesvirus Rta Lytic Switch Protein by Conditional Nuclear Localization". Journal of Virology. 82 (21): 10709–23. doi:10.1128/JVI.01012-08. PMC 2573185. PMID 18715905.
- ^ Zhang, Tiejun; Wang, Linding (2016). "Epidemiology of Kaposi's sarcoma-associated Herpesvirus in Asia: Challenges and Opportunities". Journal of Medical Virology. doi:10.1002/jmv.24662. ISSN 0146-6615.
- ^ a b Chatlynne, L. G.; Ablashi, D. V. (1999). "Seroepidemiology of Kaposi's sarcoma-associated herpesvirus (KSHV)". Seminars in Cancer Biology. 9 (3): 175–85. doi:10.1006/scbi.1998.0089. PMID 10343069.
- ^ Mohanna, S; Maco, V; Bravo, F; Gotuzzo, E (2005). "Epidemiology and clinical characteristics of classic Kaposi's sarcoma, seroprevalence, and variants of human herpesvirus 8 in South America: A critical review of an old disease". International Journal of Infectious Diseases. 9 (5): 239–50. doi:10.1016/j.ijid.2005.02.004. PMID 16095940.
- ^ a b c Fu, B; Sun, F; Li, B; Yang, L; Zeng, Y; Sun, X; Xu, F; Rayner, S; Guadalupe, M; Gao, S. J.; Wang, L (2009). "Seroprevalence of Kaposi's sarcoma-associated herpesvirus and risk factors in Xinjiang, China". Journal of Medical Virology. 81 (8): 1422–31. doi:10.1002/jmv.21550. PMC 2755560. PMID 19551832.
- ^ a b Pelser, C; Vitale, F; Whitby, D; Graubard, B. I.; Messina, A; Gafà, L; Brown, E. E.; Anderson, L. A.; Romano, N; Lauria, C; Goedert, J. J. (2009). "Socio-economic and other correlates of Kaposi sarcoma-associated herpesvirus seroprevalence among older adults in Sicily". Journal of Medical Virology. 81 (11): 1938–44. doi:10.1002/jmv.21589. PMC 2784645. PMID 19777527.
- ^ a b Wang, H; Liu, J; Dilimulati; Li, L; Ren, Z; Wen, H; Wang, X (2011). "Seroprevalence and risk factors of Kaposi's sarcoma-associated herpesvirus infection among the general Uygur population from south and north region of Xinjiang, China". Virology Journal. 8: 539. doi:10.1186/1743-422X-8-539. PMC 3266657. PMID 22168313.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Olsen, S. J.; Chang, Y; Moore, P. S.; Biggar, R. J.; Melbye, M (1998). "Increasing Kaposi's sarcoma-associated herpesvirus seroprevalence with age in a highly Kaposi's sarcoma endemic region, Zambia in 1985". AIDS (London, England). 12 (14): 1921–5. doi:10.1097/00002030-199814000-00024. PMID 9792393.
- ^ Wawer, M. J.; Eng, S. M.; Serwadda, D; Sewankambo, N. K.; Kiwanuka, N; Li, C; Gray, R. H. (2001). "Prevalence of Kaposi sarcoma-associated herpesvirus compared with selected sexually transmitted diseases in adolescents and young adults in rural Rakai District, Uganda". Sexually transmitted diseases. 28 (2): 77–81. doi:10.1097/00007435-200102000-00003. PMID 11234789.
- ^ Klaskala, W; Brayfield, B. P.; Kankasa, C; Bhat, G; West, J. T.; Mitchell, C. D.; Wood, C (2005). "Epidemiological characteristics of human herpesvirus-8 infection in a large population of antenatal women in Zambia". Journal of Medical Virology. 75 (1): 93–100. doi:10.1002/jmv.20242. PMID 15543582.
- ^ a b Minhas, V; Crabtree, K. L.; Chao, A; m'Soka, T. J.; Kankasa, C; Bulterys, M; Mitchell, C. D.; Wood, C (2008). "Early childhood infection by human herpesvirus 8 in Zambia and the role of human immunodeficiency virus type 1 coinfection in a highly endemic area". American Journal of Epidemiology. 168 (3): 311–20. doi:10.1093/aje/kwn125. PMC 2727264. PMID 18515794.
- ^ Kourí, V; Eng, S. M.; Rodríguez, M. E.; Resik, S; Orraca, O; Moore, P. S.; Chang, Y (2004). "Seroprevalence of Kaposi's sarcoma-associated herpesvirus in various populations in Cuba". Revista panamericana de salud publica = Pan American journal of public health. 15 (5): 320–5. doi:10.1590/s1020-49892004000500006. PMID 15231079.
- ^ Schulz, T. F. (2000). "Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8): Epidemiology and pathogenesis". The Journal of antimicrobial chemotherapy. 45 Suppl T3: 15–27. doi:10.1093/jac/45.suppl_4.15. PMID 10855768.
- ^ Brayfield, B. P.; Phiri, S; Kankasa, C; Muyanga, J; Mantina, H; Kwenda, G; West, J. T.; Bhat, G; Marx, D. B.; Klaskala, W; Mitchell, C. D.; Wood, C (2003). "Postnatal human herpesvirus 8 and human immunodeficiency virus type 1 infection in mothers and infants from Zambia". The Journal of Infectious Diseases. 187 (4): 559–68. doi:10.1086/367985. PMID 12599072.
- ^ He, J; Bhat, G; Kankasa, C; Chintu, C; Mitchell, C; Duan, W; Wood, C (1998). "Seroprevalence of human herpesvirus 8 among Zambian women of childbearing age without Kaposi's sarcoma (KS) and mother-child pairs with KS". The Journal of Infectious Diseases. 178 (6): 1787–90. doi:10.1086/314512. PMID 9815235.
- ^ Liu Z, Fang Q, Zuo J, Minhas V, Wood C, He N, Zhang T (2017) Was Kaposi's sarcoma-associated herpesvirus introduced into China via the ancient Silk Road? An evolutionary perspective. Arch Virol
- ^ Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA (April 1999). "Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group". The New England Journal of Medicine. 340 (14): 1063–70. doi:10.1056/NEJM199904083401402. PMID 10194235.
- ^ Yarchoan, R.; Tosato, G.; Little, R. F. (2005). "Therapy insight: AIDS-related malignancies--the influence of antiviral therapy on pathogenesis and management". Nature Clinical Practice Oncology. 2 (8): 406–415, quiz 415. doi:10.1038/ncponc0253. PMID 16130937.
Further reading
- Edelman DC (2005). "Human herpesvirus 8 – A novel human pathogen". Virology Journal. 2: 78. doi:10.1186/1743-422X-2-78. PMC 1243244. PMID 16138925.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
External links
- Human Herpesvirus-8: Related Resources HIV InSite
