Pyridoxine 5′-phosphate oxidase
| Pyridoxal 5′-phosphate synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 1.4.3.5 | ||||||||
| CAS no. | 9029-21-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Pyridoxine 5′-phosphate oxidase is an enzyme, encoded by the PNPO gene,[1][2][3] that catalyzes several reactions in the vitamin B6 metabolism pathway. Pyridoxine 5′-phosphate oxidase catalyzes the final, rate-limiting step in vitamin B6 metabolism, the biosynthesis of pyridoxal 5′-phosphate, the biologically active form of vitamin B6 which acts as an essential cofactor.[4] Pyridoxine 5′-phosphate oxidase is a member of the enzyme class oxidases, or more specifically, oxidoreductases. These enzymes catalyze a simultaneous oxidation-reduction reaction. The substrate oxidase enzymes is hydroxylated by one oxygen atom of molecular oxygen.[5] Concurrently, the other oxygen atom is reduced to water. Even though molecular oxygen is the electron acceptor in these enzymes' reactions, they are unique because oxygen does not appear in the oxidized product.
The active form of vitamin B6, pyridoxal 5'-phosphate (PLP), is critical for normal cellular function. Some cancer cells have notable differences in vitamin B6 metabolism compared to their normal counterparts. The rate-limiting enzyme in vitamin B6 synthesis is pyridoxine-5'-phosphate oxidase (PNPO; EC 1.4.3.5).[supplied by OMIM][3]
Structure
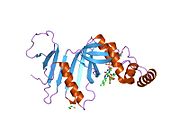
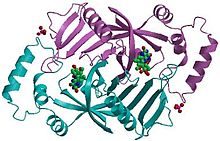
[edit]Pyridoxine 5′-phosphate oxidase is a homodimer, or a molecule consisting of two identical polypeptide subunits. It is hypothesized that the two monomers are held together by disulfide bonds. There are also salt-bridge interactions between the two monomers. Each subunit tightly binds one molecule of pyridoxal 5′-phosphate. Both alpha-helices and beta-sheets are present in the protein motif, which is best described as a split-barrel structure. This structure is due, in part, to the disulfide bonds present in the secondary protein structure of this enzyme. Multiple thiol groups (–SH) indicate the presence of disulfide bonds in the structure of the molecule. This enzyme requires the presence of a cofactor, FMN (flavin mononucleotide).[6] Cofactors are ions or coenzymes necessary for enzyme activity. The FMN is located in a deep cleft (formed by the two polypeptide subunits), and held in place by extensive hydrogen-bond interactions with the protein. In this particular case, the FMN helps the enzyme to bind the substrates. In the absence of pyridoxal 5′-phosphate (PLP), the active site of the enzyme is in an “open” conformation. Once substrate binds and is converted to PLP, the active site of the enzyme is in a partially “closed” conformation. Specific amino acid residues can form hydrogen bonds with the PLP, thus forming a lid that physically covers the active site, giving rise to the “closed” conformation.[7]

Pathway
[edit]Pyridoxine 5′-phosphate oxidase is the enzyme that catalyzes the rate-limited step of the B6 metabolism pathway. Vitamin B6, which is also known as pyridoxine, is a crucial nutrient for the human body, as it is responsible for more bodily functions than any other vitamin. Vitamin B6 is a coenzyme in the metabolism of carbohydrates, fats and proteins. This means that the enzymes which break these entities down for use in the body cannot function unless Vitamin B6 is present to induce a conformational change in the enzyme, thus activating it. Vitamin B6 also plays a role in the synthesis of hormones, red blood cells, neurotransmitters and enzymes. A person who is deficient in Vitamin B6 could suffer from insomnia, as well as suffer damage to the central nervous system.[4]
Reactions
[edit]Pyridoxine 5′-phosphate oxidase catalyzes several reactions; the two most important are the deamination of pyridoxamine 5′-phosphate and the deamination of pyridoxine 5-phosphate, both of which are key intermediates in the metabolism of B6.[8] Pyridoxine 5′-phosphate oxidase's EC number is 1.4.3.5.[6]

- pyridoxine phosphate + O2 ⇌ H2O2 + pyridoxal phosphate

Pyridoxine 5′-phosphate oxidase also plays a role in nitrogen metabolism, converting amines to aldehydes and NH3 by the reaction:
- amine + H2O + O2 ⇌ aldehyde + NH3 + H2O2

Kinetics
[edit]In humans, the pyridoxine 5′-phosphate oxidase enzyme exhibits a low catalytic rate constant of 0.2 s−1, with low Km values for both pyridoxine 5′-phosphate and pyridoxamine 5′-phosphate. The enzyme also has a low turnover rate, meaning that it is relatively slow converting substrate to product. Pyridoxal 5′-phosphate is an effective product inhibitor. Since pyridoxal 5′-phosphate, the active form of vitamin B6, is the product of the metabolic pathway, if it exists in excess, then the pathway need not proceed to keep making product. However, if it exists in low concentrations, then that is a signal for the pathway to synthesize more. This is an example of feedback inhibition.[9]
Pyridoxine 5′-phosphate oxidase in different organisms
[edit]Pyridoxine 5′-phosphate oxidase has been highly conserved over time, as there are many similarities between the enzyme as it is found in humans and Escherichia coli. Although there is only 39% retention of amino acid sequence from the E. coli version of the enzyme to the human version, the sequences for the FMN binding site and the substrate active sites are among the very highly conserved portion. One of the key differences is that the human pyridoxine 5′-phosphate oxidase has a higher specificity for the pyridoxamine 5′-phosphate substrate, whereas the pyridoxine 5′-phosphate oxidase in E. coli has a higher specificity pyridoxal 5′-phosphate substrate.[9]
Clinical significance
[edit]Mutations of the PNPO gene may result in the development of pyridoxamine 5'-phosphate oxidase deficiency, a disease presenting soon after birth with seizures and subsequent encephalopathy.
References
[edit]- ^ Ngo EO, LePage GR, Thanassi JW, Meisler N, Nutter LM (Jun 1998). "Absence of pyridoxine-5'-phosphate oxidase (PNPO) activity in neoplastic cells: isolation, characterization, and expression of PNPO cDNA". Biochemistry. 37 (21): 7741–8. doi:10.1021/bi972983r. PMID 9601034.
- ^ Kang JH, Hong ML, Kim DW, Park J, Kang TC, Won MH, Baek NI, Moon BJ, Choi SY, Kwon OS (Jun 2004). "Genomic organization, tissue distribution and deletion mutation of human pyridoxine 5'-phosphate oxidase". Eur J Biochem. 271 (12): 2452–61. doi:10.1111/j.1432-1033.2004.04175.x. PMID 15182361.
- ^ a b "Entrez Gene: PNPO pyridoxamine 5'-phosphate oxidase".
- ^ a b "Vitamin B6". Retrieved 2007-06-03.
- ^ Nelson DL, Cox MM (2005). Lehninger Principles of Biochemistry, Fourth Edition. New York: W. H. Freeman and Company. ISBN 0-7167-4339-6.
- ^ a b Online Mendelian Inheritance in Man (OMIM): Pyridoxamine 5-Prime-Phosphate Oxidase; PNPO - 603287
- ^ PDB: 1jnw; di Salvo ML, Ko TP, Musayev FN, Raboni S, Schirch V, Safo MK (January 2002). "Active site structure and stereospecificity of Escherichia coli pyridoxine-5'-phosphate oxidase". Journal of Molecular Biology. 315 (3): 385–97. doi:10.1006/jmbi.2001.5254. PMID 11786019.
- ^ "Vitamin B6 metabolism". Reference pathway. KEGG: Kyoto Encyclopedia of Genes and Genomes.
- ^ a b Musayev FN, Di Salvo ML, Ko TP, Schirch V, Safo MK (July 2003). "Structure and properties of recombinant human pyridoxine 5'-phosphate oxidase". Protein Science. 12 (7): 1455–63. doi:10.1110/ps.0356203. PMC 2323923. PMID 12824491.
Further reading
[edit]- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Musayev FN, Di Salvo ML, Ko TP, et al. (2004). "Structure and properties of recombinant human pyridoxine 5′-phosphate oxidase". Protein Sci. 12 (7): 1455–63. doi:10.1110/ps.0356203. PMC 2323923. PMID 12824491.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Mills PB, Surtees RA, Champion MP, et al. (2005). "Neonatal epileptic encephalopathy caused by mutations in the PNPO gene encoding pyridox(am)ine 5'-phosphate oxidase". Hum. Mol. Genet. 14 (8): 1077–86. doi:10.1093/hmg/ddi120. PMID 15772097.
- Kimura K, Wakamatsu A, Suzuki Y, et al. (2006). "Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes". Genome Res. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
External links
[edit] Media related to Pyridoxine 5'-phosphate oxidase at Wikimedia Commons
Media related to Pyridoxine 5'-phosphate oxidase at Wikimedia Commons