Alternative splicing: Difference between revisions
| Line 7: | Line 7: | ||
* '''Alternative donor/acceptor site mode''': in this case, alternative 5' and 3' sites are used for a given exon resulting in frame shift or insertion/deletion of amino acids in the expressed protein. |
* '''Alternative donor/acceptor site mode''': in this case, alternative 5' and 3' sites are used for a given exon resulting in frame shift or insertion/deletion of amino acids in the expressed protein. |
||
[[Image:Hyaluronidase3.gif|right|thumb|220px|Cutoff from 3 splicing structures in the murine hyaluronidase gene]]However, the terminology of these traditional "modes" may be ambigous considering complex splicing evidence. The figure to the right shows 3 spliceforms from the murine hyaluronidase 3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows the classical "intron retaining mode", whereas the comparison between the first and the third spliceform (green vs. blue) exhibits an "alternative donor site mode". A model to overcome this limitation has been proposed in <ref> |
[[Image:Hyaluronidase3.gif|right|thumb|220px|Cutoff from 3 splicing structures in the murine hyaluronidase gene]]However, the terminology of these traditional "modes" may be ambigous considering complex splicing evidence. The figure to the right shows 3 spliceforms from the murine hyaluronidase 3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows the classical "intron retaining mode", whereas the comparison between the first and the third spliceform (green vs. blue) exhibits an "alternative donor site mode". A model to overcome this limitation has been proposed in <ref> |
||
{{cite journal| |
|||
title=A general definition and nomenclature for alternative splicing events| |
|||
author=Michael Sammeth| |
|||
''PLoS Comput Biol.'' '''4''': e1000147. doi:10.1371/journal.pcbi.1000147.</ref>. |
|||
coauthors=Sylvain Foissac; Roderic Guigó| |
|||
journal=PLoS Comput Biol.| |
|||
date=8 August, 2008| |
|||
doi=10.1371/journal.pcbi.1000147| |
|||
volume=4| |
|||
pages=e1000147| |
|||
url=http://www.ploscompbiol.org/article/info:doi%2F10.1371%2Fjournal.pcbi.1000147 |
|||
}}</ref>. |
|||
== Splicing mechanism == |
== Splicing mechanism == |
||
Revision as of 14:17, 28 September 2008
Alternative splicing is the RNA splicing variation mechanism in which the exons of the primary gene transcript, the pre-mRNA, are separated and reconnected so as to produce alternative ribonucleotide arrangements. These linear combinations then undergo the process of translation where specific and unique sequences of amino acids are specified, resulting in isoform proteins. In this way, alternative splicing uses genetic expression to facilitate the synthesis of a greater variety of proteins.
Modes
There are several modes of alternative splicing, including:
- Intron retaining mode: in this case, instead of splicing out an intron, the intron is retained in the mRNA transcript. However, the intron must be properly encoding for amino acids. The intron's code must be properly expressible, otherwise a stop codon or a shift in the reading frame will cause the protein to be non-functional.
- Exon cassette mode: in this case, certain exons are spliced out to alter the sequence of amino acids in the expressed protein.
- Alternative donor/acceptor site mode: in this case, alternative 5' and 3' sites are used for a given exon resulting in frame shift or insertion/deletion of amino acids in the expressed protein.
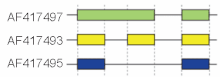
However, the terminology of these traditional "modes" may be ambigous considering complex splicing evidence. The figure to the right shows 3 spliceforms from the murine hyaluronidase 3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows the classical "intron retaining mode", whereas the comparison between the first and the third spliceform (green vs. blue) exhibits an "alternative donor site mode". A model to overcome this limitation has been proposed in [1].
Splicing mechanism
When the pre-mRNA has been transcribed from the DNA, it includes several introns and exons. (In nematodes, the mean is 4-5 exons and introns; in the fruit fly Drosophila there can be more than 100 introns and exons in one transcribed pre-mRNA.) The exons which are retained in the mRNA are determined during the splicing process. The regulation and selection of splice sites is done by Serine/Arginine-residue proteins, or SR proteins.
The intron consists of GU at 5' end and AG at 3' end, with a branch site (A) in the middle and a (py)n, denoting the polypyrimidine tract prior to the 3' end. When splicing starts, the branch site A attacks the 5' end G to form a 2',5'-phosphodiester linkage. Then the 3' end of upstream exon (G) captures the 3' end of intron by forming phosphodiester bond again, so that two exons are joined together, leaving a free intron in lariat form. In mRNA splicing, snRNPs are involved, namely, U1 to U6. For example, when splicing mRNA, U1 binds to 5' GU and U2 binds to branch site (A), then U4,U5,U6 complex comes, and U6 replaces the U1 position. U1 and U4 leaves, then U2 and U6 associate to form the lariat intron, and U5 helps bring the upstream and downstream exons together. U3 is not involved in mRNA splicing.
Importance in molecular genetics
Alternative splicing is of great importance to genetics - it invalidates the old theory of one DNA sequence coding for one polypeptide (the "one-gene-one-protein" hypothesis). External information is needed in order to decide which polypeptide is produced, given a DNA sequence and pre-mRNA. (This does not necessarily negate the central dogma of molecular biology which is about the flow of information from genes to proteins). Since the methods of regulation are inherited, the interpretation of a mutation may be changed.
It has been proposed that for eukaryotes it was a very important step towards higher efficiency, because information can be stored much more economically. Several proteins can be encoded in a DNA sequence whose length would only be enough for two proteins in the prokaryote way of coding. Others have noted that it is unnecessary to change the DNA of a gene for the evolution of a new protein. Instead, a new way of regulation could lead to the same effect, but leaving the code for the established proteins unharmed.
Another speculation is that new proteins could be allowed to evolve much faster than in prokaryotes. Furthermore, they are based on hitherto functional amino acid subchains. This may allow for a higher probability for a functional new protein. Therefore the adaptation to new environments can be much faster - with fewer generations - than in prokaryotes. This might have been one very important step for multicellular organisms with a longer life cycle.
A common myth is that alternative splicing is responsible for humans supposedly being the most complex animals, saying that humans perform more alternative splicing than the other animals. However, this is not the case. A study conducted on the subject found that "the amount of alternative splicing is comparable, with no large differences between humans and other animals."[2] The "record-holder" for alternative splicing is actually a Drosophila gene called Dscam, which has 38 016 splice variants.
References
- ^
Michael Sammeth (8 August, 2008). "A general definition and nomenclature for alternative splicing events". PLoS Comput Biol. 4: e1000147. doi:10.1371/journal.pcbi.1000147.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: unflagged free DOI (link) - ^
David Brett (17 December, 2001). "Alternative splicing and genome complexity". Nature Genetics. 30: 29–30. doi:10.1038/ng803.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help)
External links
- AStalavista (Alternative Splicing landscape visualization tool), a method for the computationally exhaustive classification of Alternative Splicing Structures
- HHMI article on alternative splicing
- Study on alternative splicing and complexity
- Stamms-lab.net: Research Group dealing with alternative Splicing issues and mis-splicing in human diseases
- Alternative Splicing of ion channels in the brain, connected to mental and neurological diseases
- BIPASS: Web Services in Alternative Splicing
- Black: Mechanisms of alternative pre-messenger RNA splicing. (review on alternative splicing mechanisms)
