Isocitrate lyase
| Isocitrate Lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Homotetrameric structure of Isocitrate lyase from E. coli. Based on PDB 1IGW.[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.3.1 | ||||||||
| CAS no. | 9045-78-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Isocitrate lyase family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ICL | ||||||||
| Pfam | PF00463 | ||||||||
| InterPro | IPR000918 | ||||||||
| PROSITE | PDOC00145 | ||||||||
| SCOP2 | 1f8m / SCOPe / SUPFAM | ||||||||
| |||||||||
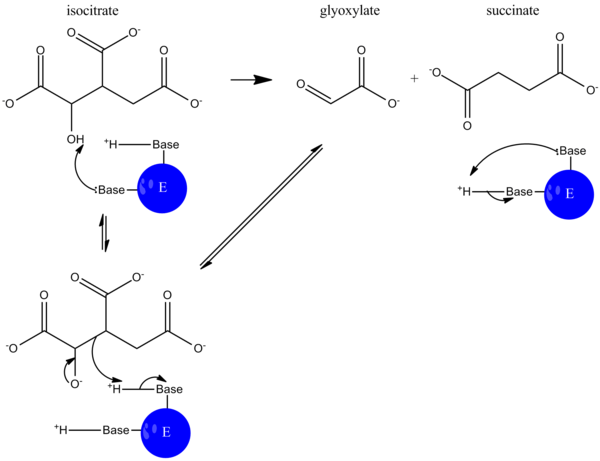
Isocitrate lyase (EC 4.1.3.1), or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate.[2][3] Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle (TCA cycle) and is used by bacteria, fungi, and plants.[4]
The systematic name of this enzyme class is isocitrate glyoxylate-lyase (succinate-forming). Other names in common use include isocitrase, isocitritase, isocitratase, threo-Ds-isocitrate glyoxylate-lyase, and isocitrate glyoxylate-lyase. This enzyme participates in glyoxylate and dicarboxylate metabolism.
Mechanism[edit]
This enzyme belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. Other enzymes also belong to this family including carboxyvinyl-carboxyphosphonate phosphorylmutase (EC 2.7.8.23) which catalyses the conversion of 1-carboxyvinyl carboxyphosphonate to 3-(hydrohydroxyphosphoryl) pyruvate carbon dioxide, and phosphoenolpyruvate mutase (EC 5.4.2.9), which is involved in the biosynthesis of phosphinothricin tripeptide antibiotics.
During catalysis, isocitrate is deprotonated, and an aldol cleavage results in the release of succinate and glyoxylate. This reaction mechanism functions much like that of aldolase in glycolysis, where a carbon-carbon bond is cleaved and an aldehyde is released.[5]
In the glyoxylate cycle, malate synthase then catalyzes the condensation of glyoxylate and acetyl-CoA to form malate so the cycle can continue.
ICL competes with isocitrate dehydrogenase, an enzyme found in the TCA cycle, for isocitrate processing. Flux through these enzymes is controlled by phosphorylation of isocitrate dehydrogenase, which has a much higher affinity for isocitrate as compared to ICL.[6] Deactivation of isocitrate dehydrogenase by phosphorylation thus leads to increased isocitrate channeling through ICL, as seen when bacteria are grown on acetate, a two-carbon compound.[6]
Enzyme structure[edit]
As of 2023, multiple structures of ICL have been solved. These include one structure from Pseudomonas aeruginosa (PDB accession code 6G1O), one structure from Fusarium graminearum (5E9H), one structure from fungus Aspergillus nidulans (1DQU), one structure from Yersinia pestis (3LG3), one structure from Burkholderia pseudomallei (3I4E), one structure from Escherichia coli (1IGW), two structures from Magnaporthe oryzae (5E9F and 5E9G), four structures from Brucella melitensis (3P0X, 3OQ8, 3EOL and 3E5B) and eleven structures from Mycobacterium tuberculosis (1F61, 1F8I, 1F8M, 6C4A, 6C4C, 5DQL, 6EDW, 6EDZ, 6EE1, 6XPP and 8G8K).
ICL is composed of four identical chains and requires a Mg2+ or Mn2+ and a thiol for activity.[4] In Escherichia coli, Lys-193, Lys-194, Cys-195, His-197, and His-356 are thought to be catalytic residues, while His-184 is thought to be involved in the assembly of the tetrameric enzyme.[7]
Between prokaryotes and eukaryotes, a difference in ICL structure is the addition of approximately 100 amino acids near the center of the eukaryotic enzyme. In eukaryotes, the additional amino acids are thought to function in the localization of ICL to single-membrane-bound organelles called glyoxysomes.[4][8] These additional amino acids account for the difference in molecular mass: the prokaryotic ICL is 48kDa, while the eukaryotic ICL is 67 kDa.[4] Only one cysteine residue is conserved between the sequences of the fungal, plant and bacterial enzymes; it is located in the middle of a conserved hexapeptide.
Most ICLs that have been characterised to date contain only one domain (the catalytic domain). However, in the isoform 2 of M. tuberculosis ICL, two domains were found.[9] Through structural and kinetic studies, the C-terminal domain was found to be a regulatory domain, which dimerises with the corresponding C-terminal domain from another subunit (of the ICL2 tetramer) upon the binding of acetyl coenzyme A to activate the catalytic activity of the enzyme.[9]
In M. tuberculosis H37Rv (a commonly-used laboratory strain), the gene that encodes ICL2 was split into two open reading frames (rv1915 and rv1916), thus encoding Rv1915 (ICL2a) and Rv1916 (ICL2b) respectively. The biological functions of Rv1915 (ICL2a) and Rv1916 (ICL2b) are poorly understood. Rv1915 and rv1916 were initially characterized as pseudogenes.[10] An in silico study in 2019 predicted that Rv1916 (ICL2b) could be involved in the synthesis of secondary metabolites.[11] In vitro studies showed that both Rv1915 (ICL2a) and Rv1916 (ICL2b) may be able to catalyze the conversion of isocitrate to form succinate and glyoxylate.[11][12] However, a recent structural and biochemical study showed that Rv1916 (ICL2b) does not have ICL activity.[13] Instead, the study showed that Rv1916 (ICL2b) is a acetyl-CoA-binding protein with unknown biological function.[13]
Assays[edit]
Several assays were developed to study the enzyme kinetics and inhibition of ICL. The most frequently-used assays involved the use of chemical or enzyme-coupled ultraviolet–visible (UV/vis) spectroscopy to measure the amount of glyoxylate that is being formed. For example, glyoxylate can be reacted with phenylhydrazine to form hydrazone that can be analysed by UV/vis spectroscopy.[14] Alternatively, lactate dehydrogenase can be used to catalyse the reduction of glyoxylate to glycolate in the presence of nicotinamide adenine dinucleotide (NADH), which is a cosubstrate of lactate dehydrogenase. During the reaction, NADH is being oxidised to NAD+. The decrease in NADH concentration can then measured by UV/vis spectroscopy using a dye.[10] In additional to spectroscopic techniques, biophysical techniques including native non-denaturing mass spectrometry and nuclear magnetic resonance (NMR) spectroscopy have also been applied to study ICL.[15][16]
Biological function[edit]
The ICL enzyme has been found to be functional in various archaea, bacteria, protists, plants, fungi, and nematodes.[17] Although the gene has been found in genomes of nematodes and cnidaria, it has not been found in the genomes of placental mammals.[17]
By diverting isocitrate from the TCA cycle, the actions of ICL and malate synthase in the glyoxylate cycle result in the net assimilation of carbon from 2-carbon compounds.[18] Thus, while the TCA cycle yields no net carbon assimilation, the glyoxylate cycle generates intermediates that can be used to synthesize glucose (via gluconeogenesis), plus other biosynthetic products. As a result, organisms that use ICL and malate synthase are able to synthesize glucose and its metabolic intermediates from acetyl-CoA derived from acetate or from the degradation of ethanol, fatty acids, or poly-β-hydroxybutyrate.[4] This function is especially important for higher plants when using seed oils. In germinating seeds, the breakdown of oils generates acetyl-CoA. This serves as a substrate for the glyoxylate cycle, which generates intermediates which serve as a primary nutrient source prior to the beginning of production of sugars by photosynthesis.[8]
In M. tuberculosis, ICL isoforms 1 and 2 also play the role of methylisocitrate lyase, converting methylisocitrate into succinate and pyruvate.[9][19] This is important because the methylcitrate cycle is key for the survival of the bacteria on odd-chain fatty acids.[20]
Disease relevance[edit]
ICL has found to be important in human, animal, and plant pathogenesis.[4] For several agricultural crops including cereals, cucumbers, and melons, increased expression of the gene encoding ICL is important for fungal virulence.[4] For instance, increased gene expression of icl1 has been seen in the fungus Leptosphaeria maculans upon infection of canola. Inactivation of the icl1 gene leads to reduced pathogenicity of the fungus, which is thought to be a result of the inability of the fungus to use carbon sources provided by the plant.[21]
Additionally, upregulation of the glyoxylate cycle has been seen for pathogens that attack humans. This is the case for fungi such as Candida albicans, which inhabits the skin, mouth, GI tract, gut and vagina of mammals and can lead to systemic infections of immunocompromised patients; as well as for the bacterium Mycobacterium tuberculosis, the major causative agent of tuberculosis.[22][23] In this latter case, ICL has been found to be essential for survival in the host.[24] Thus, ICL is a current inhibition target for therapeutic treatments of tuberculosis.[25]
Because of its use by pathogenic fungi and bacteria, specific inhibitors are being sought for ICL and malate synthase.[4] Although some inhibitors have already been identified, including itaconate, itaconic anhydride, bromopyruvate, nitropropionate, oxalate, and malate, these are non-specific and would also inhibit other enzymes essential for host function.[4][26][27] More research is needed to identify inhibitors that selectively target enzymes in the glyoxylate cycle.
See also[edit]
References[edit]
- ^ Britton KL, Abeysinghe IS, Baker PJ, Barynin V, Diehl P, Langridge SJ, et al. (September 2001). "The structure and domain organization of Escherichia coli isocitrate lyase". Acta Crystallographica. Section D, Biological Crystallography. 57 (Pt 9): 1209–18. doi:10.1107/S0907444901008642. PMID 11526312.
- ^ Beeching JR (December 1989). "High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis". Protein Sequences & Data Analysis. 2 (6): 463–6. PMID 2696959.
- ^ Atomi H, Ueda M, Hikida M, Hishida T, Teranishi Y, Tanaka A (February 1990). "Peroxisomal isocitrate lyase of the n-alkane-assimilating yeast Candida tropicalis: gene analysis and characterization". Journal of Biochemistry. 107 (2): 262–6. doi:10.1093/oxfordjournals.jbchem.a123036. PMID 2361956.
- ^ a b c d e f g h i Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I (October 2009). "Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis". Microbiology. 155 (Pt 10): 3166–75. doi:10.1099/mic.0.030858-0. PMID 19684068.
- ^ Garrett R, Grisham CN (2008). Biochemistry. Brooks Cole. pp. 588. ISBN 978-0-495-10935-8.
- ^ a b Cozzone AJ (1998). "Regulation of acetate metabolism by protein phosphorylation in enteric bacteria". Annual Review of Microbiology. 52: 127–64. doi:10.1146/annurev.micro.52.1.127. PMID 9891796.
- ^ Rehman A, McFadden BA (July 1997). "Lysine 194 is functional in isocitrate lyase from Escherichia coli". Current Microbiology. 35 (1): 14–7. doi:10.1007/s002849900203. PMID 9175553. S2CID 23972776.
- ^ a b Eastmond PJ, Graham IA (February 2001). "Re-examining the role of the glyoxylate cycle in oilseeds". Trends in Plant Science. 6 (2): 72–8. doi:10.1016/S1360-1385(00)01835-5. PMID 11173291.
- ^ a b c Bhusal, R. P.; Jiao, W.; Kwai, B. X. C.; Reynisson, J.; Collins, A. J.; Sperry, J.; Bashiri, G.; Leung, I. K. H. (Oct 2019). "Acetyl-CoA-Mediated Activation of Mycobacterium tuberculosis Isocitrate Lyase 2". Nature Communications. 10 (1): 4639. Bibcode:2019NatCo..10.4639B. doi:10.1038/s41467-019-12614-7. PMC 6788997. PMID 31604954.
- ^ a b Höner Zu Bentrup K, Miczak A, Swenson DL, Russell DG (December 1999). "Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis". Journal of Bacteriology. 181 (23): 7161–7. doi:10.1128/JB.181.23.7161-7167.1999. PMC 103675. PMID 10572116.
- ^ a b Antil M, Sharma J, Brissonnet Y, Choudhary M, Gouin S, Gupta V (September 2019). "Structure function insights into elusive Mycobacterium tuberculosis protein Rv1916" (PDF). International Journal of Biological Macromolecules. 141: 927–936. doi:10.1016/j.ijbiomac.2019.09.038. PMID 31505209. S2CID 202556684.
- ^ Antil M, Gupta V (June 2022). "Rv1915 and Rv1916 from Mycobacterium tuberculosis H37Rv form in vitro protein-protein complex". Biochimica et Biophysica Acta (BBA) - General Subjects. 1866 (6): 130130. doi:10.1016/j.bbagen.2022.130130. PMID 35307510.
- ^ a b Huang EY, Kwai BX, Bhusal RP, Bashiri G, Leung, IK (May 2023). "Mycobacterium tuberculosis Rv1916 is an Acetyl-CoA-Binding Protein". ChemBioChem. 24 (14): e202300162. doi:10.1002/cbic.202300162. PMID 37211532.
- ^ "Proceedings of the Biochemical Society". The Biochemical Journal. 72 (1): 1P–13P. May 1959. doi:10.1042/bj0720001P. PMC 1196904. PMID 16748793.
- ^ Pham TV, Murkin AS, Moynihan MM, Harris L, Tyler PC, Shetty N, et al. (July 2017). "Mycobacterium tuberculosis". Proceedings of the National Academy of Sciences of the United States of America. 114 (29): 7617–7622. doi:10.1073/pnas.1706134114. PMC 5530696. PMID 28679637.
- ^ Bhusal RP, Patel K, Kwai BX, Swartjes A, Bashiri G, Reynisson J, et al. (November 2017). "Mycobacterium tuberculosis isocitrate lyase inhibitors". MedChemComm. 8 (11): 2155–2163. doi:10.1039/C7MD00456G. PMC 6072051. PMID 30108733.
- ^ a b Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN (October 2006). "Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation". Biology Direct. 1 (31): 31. doi:10.1186/1745-6150-1-31. PMC 1630690. PMID 17059607.
- ^ Kornberg HL, Krebs HA (May 1957). "Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle". Nature. 179 (4568): 988–91. Bibcode:1957Natur.179..988K. doi:10.1038/179988a0. PMID 13430766. S2CID 40858130.
- ^ Gould TA, van de Langemheen H, Muñoz-Elías EJ, McKinney JD, Sacchettini JC (August 2006). "Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis". Molecular Microbiology. 61 (4): 940–7. doi:10.1111/j.1365-2958.2006.05297.x. PMID 16879647. S2CID 26099043.
- ^ Eoh H, Rhee KY (April 2014). "Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids". Proceedings of the National Academy of Sciences of the United States of America. 111 (13): 4976–81. Bibcode:2014PNAS..111.4976E. doi:10.1073/pnas.1400390111. PMC 3977286. PMID 24639517.
- ^ Idnurm A, Howlett BJ (October 2002). "Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus)". Eukaryotic Cell. 1 (5): 719–24. doi:10.1128/EC.1.5.719-724.2002. PMC 126752. PMID 12455691.
- ^ Lorenz MC, Bender JA, Fink GR (October 2004). "Transcriptional response of Candida albicans upon internalization by macrophages". Eukaryotic Cell. 3 (5): 1076–87. doi:10.1128/EC.3.5.1076-1087.2004. PMC 522606. PMID 15470236.
- ^ Srivastava V, Jain A, Srivastava BS, Srivastava R (May 2008). "Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice". Tuberculosis. 88 (3): 171–7. doi:10.1016/j.tube.2007.10.002. PMID 18054522.
- ^ Muñoz-Elías EJ, McKinney JD (June 2005). "Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence". Nature Medicine. 11 (6): 638–44. doi:10.1038/nm1252. PMC 1464426. PMID 15895072.
- ^ Bhusal RP, Bashiri G, Kwai BX, Sperry J, Leung IK (July 2017). "Targeting isocitrate lyase for the treatment of latent tuberculosis". Drug Discovery Today. 22 (7): 1008–1016. doi:10.1016/j.drudis.2017.04.012. PMID 28458043.
- ^ Krátký M, Vinšová J (Dec 2012). "Advances in mycobacterial isocitrate lyase targeting and inhibitors". Current Medicinal Chemistry. 19 (36): 6126–37. doi:10.2174/0929867311209066126. PMID 23092127.
- ^ Lee YV, Wahab HA, Choong YS (2015). "Potential inhibitors for isocitrate lyase of Mycobacterium tuberculosis and non-M. tuberculosis: a summary". BioMed Research International. 2015: 895453. doi:10.1155/2015/895453. PMC 4306415. PMID 25649791.
Further reading[edit]
- McFadden BA, Howes WV (1963). "Crystallisation and some properties of isocitrate lyase from Pseudomonas indigofera". J. Biol. Chem. 238 (5): 1737–1742. doi:10.1016/S0021-9258(18)81129-2.
- Shiio I, Shiio T, Mcfadden BA (January 1965). "Isocitrate lyase from Pseudomonas indigofera I. Preparation, amino acid composition and molecular weight". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 96: 114–22. doi:10.1016/0005-2787(65)90615-5. PMID 14285253.
- Vickery HB (June 1962). "A suggested new nomenclature for the isomers of isocitric acid". The Journal of Biological Chemistry. 237 (6): 1739–41. doi:10.1016/S0021-9258(19)73928-3. PMID 13925783.