Signal transduction: Difference between revisions
divide sentence for clarity |
edit for clarity |
||
| Line 1: | Line 1: | ||
{{Refimprove|date=May 2011}} |
{{Refimprove|date=May 2011}} |
||
[[Image:Signal transduction v1.png|thumb|An overview of major signal transduction pathways.]] |
[[Image:Signal transduction v1.png|thumb|An overview of major signal transduction pathways.]] |
||
'''Signal transduction''' occurs when an extracellular signaling molecule activates a membrane receptor. In turn, this receptor alters intracellular molecules creating a response.<ref>Silverthorn (2007). Human Physiology. 4th ed.</ref> There are two stages in this process: 1) a signaling molecule activates a specific receptor on the cell membrane 2) causing a second messenger to continue the signal into the cell and elicit a physiological response. In either step, the signal can be amplified, |
'''Signal transduction''' occurs when an extracellular signaling molecule activates a membrane receptor. In turn, this receptor alters intracellular molecules creating a response.<ref>Silverthorn (2007). Human Physiology. 4th ed.</ref> There are two stages in this process: 1) a signaling molecule activates a specific receptor on the cell membrane 2) causing a second messenger to continue the signal into the cell and elicit a physiological response. In either step, the signal can be amplified. Thus, one signalling molecule can cause many responses.<ref name="Campbell">{{cite book|author=Reece, Jane; Campbell, Neil|title=Biology|publisher=Benjamin Cummings|location=San Francisco|year=2002|pages=|isbn=0-8053-6624-5|oclc=|doi=|accessdate=}}</ref> |
||
==History== |
==History== |
||
Revision as of 22:20, 24 October 2011
This article needs additional citations for verification. (May 2011) |
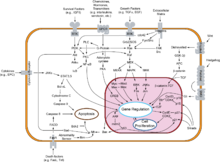
Signal transduction occurs when an extracellular signaling molecule activates a membrane receptor. In turn, this receptor alters intracellular molecules creating a response.[1] There are two stages in this process: 1) a signaling molecule activates a specific receptor on the cell membrane 2) causing a second messenger to continue the signal into the cell and elicit a physiological response. In either step, the signal can be amplified. Thus, one signalling molecule can cause many responses.[2]
History

In 1970, Martin Rodbell examined the effects of glucagon on a rat's liver cell membrane receptor. He noted that guanosine triphosphate disassociated glucagon from this receptor and stimulated the G-protein, which strongly influenced the cell's metabolism. Thus he deduced that the G-protein was a transducer that accepted glucagon molecules and affected the cell.[3] For this he shared the 1994 Nobel Prize in Physiology or Medicine with Alfred G. Gilman. The current understanding of signal transduction processes reflects contributions made by Rodbell and many other research groups.
The earliest scientific paper recorded in the MEDLINE database as containing the specific term signal transduction was published in 1972.[4] Some articles published before 1977 used the term signal transmission or sensory transduction for signal transduction:[5][6] a total of 48,377 scientific papers related to signal transduction were published in 1977, of which 11,211 were reviews of other papers. That year the actual term signal transduction was included in abstracts until in 1979 it appeared in a paper's title.[7][8] One source attributes the widespread use of this term to a 1980 review article by Rodbell:[9][3] research papers directly addressing signal transduction processes began to appear in large numbers in the late 1980s and early 1990s.
Signaling molecules
Signal transduction involves the binding of extracellular signalling molecules and ligands to cell-surface receptors that trigger events inside the cell. Intracellular signaling cascades can be started through cell-substratum interactions; examples are the integrin that binds ligands in the extracellular matrix and steroids.[10] Most steroid hormones have receptors within the cytoplasm and act by stimulating the binding of their receptors to the promoter region of steroid-responsive genes.[11] Examples of signaling molecules include the hormone melatonin,[12] the neurotransmitter acetylcholine[13] and the cytokine interferon γ.[14]
The classifications of signalling molecules do not take into account the molecular nature of each class member; neurotransmitters range in size from small molecules such as dopamine[15] to neuropeptides such as endorphins.[16] Some molecules may fit into more than one class; for example, epinephrine is a neurotransmitter when secreted by the central nervous system and a hormone when secreted by the adrenal medulla.
Environmental stimuli
With single-celled organisms, the variety of signal transduction processes influence its reaction to its environment.[citation needed] With multicellular organisms, numerous processes are required for coordinating individual cells to support the organism as a whole; the complexity of these processes tend to increase with the complexity of the organism.[citation needed] Sensing of environments at the cellular level relies on signal transduction;[citation needed] many disease processes, such as diabetes and heart disease arise from defects in these pathways, highlighting the importance of this process in biology and medicine.
Various environmental stimuli exist that initiate signal transmission processes in multicellular organisms; examples include photons hitting cells in the retina of the eye,[17] and odorants binding to odorant receptors in the nasal epithelium.[18] Certain microbial molecules, such as viral nucleotides and protein antigens, can elicit an immune system response against invading pathogens mediated by signal transduction processes. This may occur independent of signal transduction stimulation by other molecules, as is the case for the toll-like receptor. It may occur with help from stimulatory molecules located at the cell surface of other cells, as with T-cell receptor signaling. Single-celled organisms may respond to environmental stimuli through the activation of signal transduction pathways. For example, slime molds secrete cyclic adenosine monophosphate upon starvation, stimulating individual cells in the immediate environment to aggregate,[19] and yeast cells use mating factors to determine the mating types of other cells and to participate in sexual reproduction.[20]
Receptors
Receptors can be roughly divided into two major classes: intracellular receptors and extracellular receptors.
Extracellular
Extracellular receptors are integral transmembrane proteins and make up most receptors. They span the plasma membrane of the cell, with one part of the receptor on the outside of the cell and the other on the inside. Signal transduction occurs as a result of a ligand binding to the outside; the molecule does not pass through the membrane. This binding stimulates a series of events inside the cell; different types of receptor stimulate different responses and receptors typically respond to only the binding of a specific ligand. Upon binding, the ligand induces a change in the conformation of the inside part of the receptor.[21] These result in either the activation of an enzyme in the receptor or the exposure of a binding site for other intracellular signaling proteins within the cell, eventually propagating the signal through the cytoplasm.
In eukaryotic cells, most intracellular proteins activated by a ligand/receptor interaction possess an enzymatic activity; examples include tyrosine kinase and phosphatases. Some of them create second messengers such as cyclic AMP and IP3, the latter controlling the release of intracellular calcium stores into the cytoplasm. Other activated proteins interact with adapter proteins that facilitate signalling protein interactions and coordination of signalling complexes necessary to respond to a particular stimulus. Enzymes and adapter proteins are both responsive to various second messenger molecules.
Many adapter proteins and enzymes activated as part of signal transduction possess specialized protein domains that bind to specific secondary messenger molecules. For example, calcium ions bind to the EF hand domains of calmodulin, allowing it to bind and activate calmodulin-dependent kinase. PIP3 and other phosphoinositides do the same thing to the Pleckstrin homology domains of proteins such as the kinase protein AKT.
G protein-coupled
G protein-coupled receptors (GPCRs) are a family of integral transmembrane proteins that possess seven transmembrane domains and are linked to a heterotrimeric G protein. Many receptors are in this family, including adrenergic receptors and chemokine receptors.
Signal transduction by a GPCR begins with an inactive G protein coupled to the receptor; it exists as a heterotrimer consisting of Gα, Gβ, and Gγ. Once the GPCR recognizes a ligand, the conformation of the receptor changes to activate the G protein, causing Gα to bind a molecule of GTP and dissociate from the other two G-protein subunits. The dissociation exposes sites on the subunits that can interact with other molecules.[22] The activated G protein subunits detach from the receptor and initiate signalling from many downstream effector proteins such as phospholipases and ion channels, the latter permitting the release of second messenger molecules.[23] The total strength of signal amplification by a GPCR is determined by the lifetimes of the ligand-receptor complex and receptor-effector protein complex and the deactivation time of the activated receptor and effectors through intrinsic enzymatic activity.
A study was conducted where a point mutation was inserted into the gene encoding the chemokine receptor CXCR2; mutated cells underwent a malignant transformation due to the expression of CXCR2 in an active conformation despite the absence of chemokine-binding. This meant that chemokine receptors participate in cancer development.[24]
Tyrosine and histidine kinase
Receptor tyrosine kinases (RTKs) are transmembrane proteins with an intracellular kinase domain and an extracellular domain that binds ligands; examples include growth factor receptors such as the insulin receptor.[25] To perform signal transdution, RTKs need to form dimers in the plasma membrane;[26] the dimer is stabilized by ligands binding to the receptor. The interaction between the cytoplasmic domains stimulates the autophosphorylation of tyrosines within the domains of the RTKs, causing conformational changes. The receptors' kinase domains are subsequently activated, initiating phosphorylation signalling cascades of downstream cytoplasmic molecules that facilitate various cellular processes such as cell differentiation and metabolism.[25]
As is the case with GPCRs, proteins that bind GTP play a major role in signal transduction from the activated RTK into the cell. In this case, the G proteins are members of the Ras, Rho, and Raf families, referred to collectively as small G proteins. They act as molecular switches usually tethered to membranes by isoprenyl groups linked to their carboxyl ends. Upon activation, they assign proteins to specific membrane subdomains where they participate in signaling. Activated RTKs in turn activate small G proteins that activate guanine nucleotide exchange factors such as SOS1. Once activated, these exchange factors can activate more small G proteins, thus amplifying the receptor's initial signal. The mutation of certain RTK genes, as with that of GPCRs, can result in the expression of receptors that exist in a constitutively-activate state; such mutated genes may act as oncogenes.[27]
Histidine-specific protein kinases are structurally distinct from other protein kinases and are found in prokaryotes, fungi, and plants as part of a two-component signal transduction mechanism: a phosphate group from ATP is first added to a histidine residue within the kinase, then transferred to an aspartate residue on a receiver domain on a different protein or the kinase itself, thus activating the aspartate residue.[28]
Integrin

Integrins are produced by a wide variety of cells; they play a role in cell attachment to other cells and the extracellular matrix and in the transduction of signals from extracellular matrix components such as fibronectin and collagen. Ligand binding to the extracellular domain of integrins changes the protein's conformation, clustering it at the cell membrane to initiate signal transduction. Integrins lack kinase activity; hence integrin-mediated signal transduction is achieved through a variety of intracellular protein kinases and adaptor molecules, the main coordinator being integrin-linked kinase.[29] As shown in the picture to the right, cooperative integrin-RTK signalling determines the timing of cellular survival, apoptosis, proliferation, and differentiation.
Important differences exist between integrin signalling in circulating blood cells and non-circulating cells such as epithelial cells; integrins of circulating cells are normally inactive. For example, cell membrane integrins on circulating leukocytes are maintained in an inactive state to avoid epithelial cell attachment; they are only activated in response to stimuli such as those received at the site of an inflammatory response. In a similar manner, integrins at the cell membrane of circulating platelets are normally kept inactive to avoid thrombosis. Epithelial cells (which are non-circulating) normally have active integrins at their cell membrane, helping maintain their stable adhesion to underlying stromal cells that provide signals to maintain normal functioning.[30]
Toll gate
When activated, toll-like receptors (TLRs) take adapter molecules within the cytoplasm of cells in order to propagate a signal. Four adaptor molecules are known to be involved in signaling, which are Myd88, TIRAP, TRIF, and TRAM.[31][32][33] These adapters activate other intracellular molecules such as IRAK1, IRAK4, TBK1, and IKKi that amplify the signal, eventually leading to the induction or suppression of genes that cause certain responses. Thousands of genes are activated by TLR signaling, implying that this method constitutes an important gateway for gene modulation.
Ligand-gated ion channel
A ligand-gated ion channel, upon binding with a ligand, changes conformation to open a channel in the cell membrane through which ions relaying signals can pass. An example of this mechanism is found in the receiving cell of a neural synapse. The influx of ions that occurs in response to these channels opening induces action potentials, such as those that travel along nerves, by depolarizing the membrane of post-synaptic cells, resulting in the opening of voltage-gated ion channels.
An example of an ion allowed into the cell during a ligand-gated ion channel opening is Ca2+; it acts as a second messenger initiating signal transduction cascades and altering the physiology of the responding cell. This results in amplification of the synapse response between synaptic cells by remodelling the dendritic spines involved in the synapse.
Intracellular
Intracellular receptors, such as nuclear receptors and cytoplasmic receptors, are soluble proteins localized within their respective areas. The typical ligands for nuclear receptors are lipophilic hormones like the steroid hormones testosterone and progesterone and derivatives of vitamins A and D. To initiate signal transduction, the ligand must pass through the plasma membrane by passive diffusion. On binding with the receptor, the ligands pass through the nuclear membrane into the nucleus, enabling gene transcription and protein production.
Activated nuclear receptors attach to the DNA at receptor-specific hormone-responsive element (HRE) sequences, located in the promoter region of the genes activated by the hormone-receptor complex. Due to them enabling gene transcription, they are alternatively called inductors of gene expression. Activation of gene transcription is slower than signals directly affecting existing proteins; therefore, the effects of hormones that use nucleic receptors are long-term.
Signal transduction via these receptors involves little proteins, but the details of gene regulation by this method are not well understood. Nucleic receptors have DNA-binding domains containing zinc fingers and a ligand-binding domain; the zinc fingers stabilize DNA binding by holding its phosphate backbone. DNA sequences that match the receptor are usually hexameric repeats of any kind; the sequences are similar but their orientation and distance differentiate them. The ligand-binding domain is additionally responsible for dimerization of nucleic receptors prior to binding and providing structures for transactivation used for communication with the translational apparatus.
Steroid receptors are a subclass of nuclear receptors located primarily within the cytosol; in the absence of steroids, they cling together in an aporeceptor complex containing chaperone or heatshock proteins (HSPs). The HSPs are necessary to activate the receptor by assisting the protein to fold in a way such that the signal sequence enabling its passage into the nucleus is accessible. Steroid receptors, on the other hand, may be repressive on gene expression when their transactivation domain is hidden; activity can be enhanced by phosphorylation of serine residues at their N-terminal as a result of another signal transduction pathway, a process called crosstalk.
Retinoic acid receptors are another subset of nuclear receptors. They can be activated by an endocrine-synthesized ligand that entered the cell by diffusion, a ligand synthesised from a precursor like retinol brought to the cell through the bloodstream or a completely intracellularly synthesised ligand like prostaglandin. These receptors are located in the nucleus and are not accompanied by HSPs; they repress their gene by binding to their specific DNA sequence when no ligand binds to them and vice versa.
Certain intracellular receptors of the immune system are cytoplasmic receptors; recently identified NOD-like receptors (NLRs) reside in the cytoplasm of some eukaryotic cells and interact with ligands using a leucine-rich repeat (LRR) motif similar to TLRs. Some of these molecules like NOD2 interact with RIP2 kinase that activates NF-κB signaling, whereas others like NALP3 interact with inflammatory caspases and initiate processing of particular cytokines like interleukin-1β.[34]
Second messengers
Intracellular signal transduction is carried out by second messenger molecules.
Calcium
The release of calcium ions from the endoplasmic reticulum into the cytosol results in its binding to signaling proteins that are then activated; it is then sequestered in the smooth endoplasmic reticulum and the mitochondria. Two combined receptor/ion channel proteins control the transport of calcium: the InsP3-receptor that transports calcium upon interaction with inositol triphosphate on its cytosolic side and the ryanodine receptor named after the alkaloid ryanodine, similar to the InsP3 receptor but having a feedback mechanism that releases more calcium upon binding with it. The nature of calcium in the cytosol means that it is active for only a very short time, meaning its free state concentration is very low and is mostly bound to organelle molecules like calreticulin when inactive.
Calcium is used in many processes including muscle contraction, neurotransmitter release from nerve endings and cell migration. The three main pathways that lead to its activation are GPCR pathways, RTK pathways and gated ion channels; it regulates proteins either directly or by binding to an enzyme.
Lipophilics
Lipophilic second messenger molecules are derived from lipids residing in cellular membranes; enzymes stimulated by activated receptors activate the lipids by modifying them. Examples include diacylglycerol and ceramide, the former required for the activation of protein kinase C.
Nitric oxide
Nitric oxide (NO) acts as a second messenger because it is a free radical that can diffuse through the plasma membrane and affect nearby cells. It is synthesised from arginine and oxygen by the NO synthase and works through activation of soluble guanylyl cyclase, which when activated produces another second messenger, cGMP. NO can also act through covalent modification of proteins or their metal co-factors; some have a redox mechanism and are reversible. It is toxic in high concentrations and causes damage during stroke, but is the cause of many other functions like relaxation of blood vessels, apoptosis and erections.
Cellular responses
Gene activations[35] and metabolism alterations[36] are examples of cellular responses to extracellular stimulation that require signal transduction. Gene activation leads to further cellular effects, since the products of responding genes include instigators of activation; transcription factors produced as a result of a signal transduction cascade can activate even more genes. Hence, an initial stimulus can trigger the expression of a large number of genes, leading to physiological events like the increased uptake of glucose from the blood stream[36] and the migration of neutrophils to sites of infection. The set of genes and their activation order to certain stimuli is referred to as a genetic program.[37]
Mammalian cells require stimulation for cell division and survival; in the absence of growth factor, apoptosis ensues. Such requirements for extracellular stimulation are necessary for controlling cell behavior in unicellular and multicellular organisms; signal transduction pathways are perceived to be so central to biological processes that a large number of diseases are attributed to their disregulation.
Major pathways
Following are some major signaling pathways, demonstrating how ligands binding to their receptors can affect second messengers and eventually result in altered cellular responses.
- MAPK/ERK pathway: A pathway that couples intracellular responses to the binding of growth factors to cell surface receptors. This pathway is very complex and includes many protein components.[38] In many cell types, activation of this pathway promotes cell division, and many forms of cancer are associated with aberrations in it.[39]
- cAMP dependent pathway: In humans, cAMP works by activating protein kinase A (PKA, cAMP-dependent protein kinase) (see picture), and thus, further effects mainly depend on cAMP-dependent protein kinase, which vary based on the type of cell.
- IP3/DAG pathway: PLC cleaves the phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2) yielding diacyl glycerol (DAG) and inositol 1,4,5-triphosphate (IP3). DAG remains bound to the membrane, and IP3 is released as a soluble structure into the cytosol. IP3 then diffuses through the cytosol to bind to IP3 receptors, particular calcium channels in the endoplasmic reticulum (ER). These channels are specific to calcium and only allow the passage of calcium to move through. This causes the cytosolic concentration of Calcium to increase, causing a cascade of intracellular changes and activity.[40] In addition, calcium and DAG together works to activate PKC, which goes on to phosphorylate other molecules, leading to altered cellular activity. End effects include taste, manic depression, tumor promotion, etc.[40]
See also
- Two-component regulatory system
- Functional selectivity
- G protein-coupled receptor
- GTPases
- Protein phosphatase
- Redox signaling
- Transduction
References
- ^ Silverthorn (2007). Human Physiology. 4th ed.
- ^ Reece, Jane; Campbell, Neil (2002). Biology. San Francisco: Benjamin Cummings. ISBN 0-8053-6624-5.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b Rodbell, M. (1980). "The role of hormone receptors and GTP-regulatory proteins in membrane transduction". Nature. 284 (5751): 17–22. doi:10.1038/284017a0. PMID 6101906.
- ^ Rensing, L. (1972). "Periodic geophysical and biological signals as Zeitgeber and exogenous inducers in animal organisms". Int. J. Biometeorol. 16: Suppl:113–125. PMID 4621276.
{{cite journal}}: Unknown parameter|nopp=ignored (|no-pp=suggested) (help) - ^ Tonndorf J. (1975). "Davis-1961 revisited. Signal transmission in the cochlear hair cell-nerve junction". Arch. Otolaryngol. 101 (9): 528–535. PMID 169771.
- ^ Ashcroft SJ, Crossley JR, Crossley PC. (1976). "The effect of N-acylglucosamines on the biosynthesis and secretion of insulin in the rat". Biochem. J. 154 (3): 701–707. PMC 1172772. PMID 782447.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hildebrand E. (1977). "What does Halobacterium tell us about photoreception?". Biophys. Struct. Mech. 3 (1): 69–77. doi:10.1007/BF00536457. PMID 857951.
- ^ Kenny JJ, Martinez-Maza O.; et al. (1979). "Lipid synthesis: an indicator of antigen-induced signal transduction in antigen-binding cells". J. Immunol. 112 (4): 1278–1284. PMID 376714.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Gomperts, BD. (2002). Signal transduction. Academic Press. ISBN 0-12-289631-9.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Beato M, Chavez S and Truss M (1996). "Transcriptional regulation by steroid hormones". Steroids. 61 (4): 240–251. doi:10.1016/0039-128X(96)00030-X. PMID 8733009.
- ^ Hammes SR (2003). "The further redefining of steroid-mediated signaling". Proc Natl Acad Sci USA. 100 (5): 21680–2170. doi:10.1073/pnas.0530224100. PMC 151311. PMID 12606724.
- ^ Sugden D, Davidson K.; et al. (2004). "Melatonin, melatonin receptors and melanophores: a moving story". Pigment Cell Res. 17 (5): 454–460. doi:10.1111/j.1600-0749.2004.00185.x. PMID 15357831.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Kistler J, Stroud RM; et al. (1982). "Structure and function of an acetylcholine receptor". Biophys. J. 37 (1): 371–383. Bibcode:1982BpJ....37..371K. doi:10.1016/S0006-3495(82)84685-7. PMC 1329155. PMID 7055628.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Schroder; et al. (2004). "Interferon-γ an overview of signals, mechanisms and functions". Journal of Leukocyte Biology. 75 (2): 163–189. doi:10.1189/jlb.0603252. PMID 14525967.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Missale C, Nash SR.; et al. (1998). "Dopamine receptors:from structure to function". Physiol. Rev. 78 (1): 189–225. PMID 9457173.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Goldstein, A. (1976). "Opioid peptides endorphins in pituitary and brain". Science. 193 (4258): 1081–1086. doi:10.1126/science.959823. PMID 959823.
- ^ Burns ME and Arshavsky VY. (2005). "Beyond counting photons: trials and trends in vertebrate visual transduction". Neuron. 48 (3): 387–401. doi:10.1016/j.neuron.2005.10.014. PMID 16269358.
- ^ Ronnett GV and Moon C. (2002). "G proteins and olfactory signal transduction". Annu Rev Physiol. 64 (1): 189–222. doi:10.1146/annurev.physiol.64.082701.102219. PMID 11826268.
- ^ Hanna MH, Nowicki JJ and Fatone MA (1984). "Extracellular cyclic AMP (cAMP) during development of the cellular slime mold Polysphondylium violaceum: comparison of accumulation in the wild type and an aggregation-defective mutant". J Bacteriol. 157 (2): 345–349. PMID 215252.
- ^ Sprague GF Jr. (1991). "Signal transduction in yeast mating: receptors, transcription factors, and the kinase connection". Trends Genet. 7 (11–12): 393–398. doi:10.1016/0168-9525(91)90218-F. PMID 1668192.
- ^ A molecular model for receptor activation
- ^ Jeremy M. Berg, John L. Tymoczko, Lubert Stryer; Web content by Neil D. Clarke (2002). Biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-4954-8.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Yang W, Xia S (2006). "Mechanisms of regulation and function of G-protein-coupled receptor kinases". World J Gastroenterol. 12 (48): 7753–7. PMID 17203515.
- ^ Burger M, Burger, JA; et al. (1999). "Point mutation causing constitutive signaling of CXCR2 leads to transforming activity similar to Kaposi's sarcoma herpesvirus-G protein-coupled receptor". J. Immunol. 163 (4): 2017–2022. PMID 10438939.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b Li E, Hristova K (2006). "Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies". Biochemistry. 45 (20): 6241–51. doi:10.1021/bi060609y. PMID 16700535.
- ^ Schlessinger, J. (1988). "Signal transduction by allosteric receptor oligomerization". Trends Biochem Sci. 13 (11): 443–7. doi:10.1016/0968-0004(88)90219-8. PMID 3075366.
{{cite journal}}:|access-date=requires|url=(help) - ^ Roskoski, R, Jr. (2004). "The ErbB/HER receptor protein-tyrosine kinases and cancer". Biochem. Biophys. Res. Commun. 319 (1): 1–11. doi:10.1016/j.bbrc.2004.04.150. PMID 15158434.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wolanin PW, Thomason PA, Stock JB (2002). "Histidine protein kinases: key signal transducers outside the animal kingdom". Genome Biology. 3 (10): reviews3013.1–3013.8. doi:10.1186/gb-2002-3-10-reviews3013. PMC 244915. PMID 12372152.
{{cite journal}}: Unknown parameter|pmcid=ignored (|pmc=suggested) (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b Hehlgans, S. Haase, M. and Cordes, N. (2007). "Signaling via integrins: Implications for cell survival and anticancer strategies". Biochim. Biophys. Acta. 1775 (1): 163–180. doi:10.1016/j.bbcan.2006.09.001. PMID 17084981.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gilcrease MZ. (2006). "Integrin signaling in epithelial cells". Cancer Lett. 247 (1): 1–25. doi:10.1016/j.canlet.2006.03.031. PMID 16725254.
- ^ Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S (2003). "Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway". Science. 301 (5633): 640–3. doi:10.1126/science.1087262. PMID 12855817.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S (2003). "TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway". Nat Immunol. 4 (11): 1144–50. doi:10.1038/ni986. PMID 14556004.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, Takeda K, Akira S (2002). "Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4". Nature. 420 (6913): 324–9. doi:10.1038/nature01182. PMID 12447441.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Delbridge L, O'Riordan M (2007). "Innate recognition of intracellular bacteria". Curr Opin Immunol. 19 (1): 10–6. doi:10.1016/j.coi.2006.11.005. PMID 17126540.
- ^ Lalli E and Sassone-Corsi P (1994). "Signal transduction and gene regulation: the nuclear response to cAMP". J Biol Chem. 269 (26): 17359–17362. PMID 8021233.
- ^ a b Rosen O (1987). "After insulin binds". Science. 237 (4821): 1452–1458. doi:10.1126/science.2442814. PMID 2442814.
- ^ Massague J and Gomis RR (2006). "The logic of TGFbeta signaling". FEBS Lett. 580 (12): 2811–2820. doi:10.1016/j.febslet.2006.04.033. PMID 16678165.
- ^ Orton RJ, Sturm OE, Vyshemirsky V, Calder M, Gilbert DR, Kolch W (2005). "Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway". The Biochemical journal. 392 (Pt 2): 249–61. doi:10.1042/BJ20050908. PMC 1316260. PMID 16293107.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 15286780, please use {{cite journal}} with
|pmid=15286780instead. - ^ a b Alberts B, Lewis J, Raff M, Roberts K, Walter P (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. ISBN 0-8153-3218-1.
{{cite book}}: CS1 maint: multiple names: authors list (link)
External links
- Netpath - A curated resource of signal transduction pathways in humans
- Signal Transduction - The Virtual Library of Biochemistry and Cell Biology
- TRANSPATH(R) - A database about signal transduction pathways
- Redox Signaling Molecules - A public discussion.
- Redox Signaling Molecules - PDf download
- Science's STKE - Signal Transduction Knowledge Environment, from the journal Science, published by AAAS.
- Signal+Transduction at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- UCSD-Nature Signaling Gateway, from Nature Publishing Group
- LitInspector - Signal transduction pathway mining in PubMed abstracts
- Huaxian Chen, et al. A Cell Based Immunocytochemical Assay For Monitoring Kinase Signaling Pathways And Drug Efficacy (PDF) Analytical Biochemistry 338 (2005) 136-142
- www.Redoxsignaling.com
- Signaling PAthway Database - Kyushu University
- Cell cycle - Homo sapiens (human) - KEGG PATHWAY [1]
- Pathway Interaction Database - NCI
