Proline: Difference between revisions
FeatherPluma (talk | contribs) format |
→Properties in protein structure: Moved the simpler bits to the front, added an intro before the first greek letter and removed the too technical tag |
||
| Line 91: | Line 91: | ||
==Properties in protein structure== |
==Properties in protein structure== |
||
The distinctive cyclic structure of proline's side chain gives proline an exceptional conformational rigidity compared to other amino acids. It also affects the rate of peptide bond formation between proline and other amino acids. When proline is bound as an amide in a peptide bond, its nitrogen is not bound to any hydrogen, meaning it cannot act as a [[hydrogen bond]] donor, but can be a hydrogen bond acceptor. |
|||
{{Technical|section|date=September 2012}} |
|||
| ⚫ | |||
| ⚫ | Peptide bond formation with incoming Pro-tRNA<sup>Pro</sup> is considerably slower than with any other tRNAs, which is a general feature of N-alkylamino acids.<ref>{{citation | first1 = Michael Y | last1 = Pavlov | first2 = Richard E | last2 = Watts | first3 = Zhongping | last3 = Tan | first4 = Virginia W | last4 = Cornish | first5 = Måns | last5 = Ehrenberg | first6 = Anthony C | title = Slow peptide bond formation by proline and other N-alkylamino acids in translation | year = 2010 | journal = PNAS | volume = 106 | issue = 1 | pages = 50–54 | doi = 10.1073/pnas.0809211106 | pmid = 19104062 | last6 = Forster | pmc = 2629218}}.</ref> Peptide bond formation is also slow between an incoming tRNA and a chain ending in proline; with the creation of proline-proline bonds slowest of all.<ref>{{Cite journal |last=Buskirk |first=Allen R. |last2=Green |first2=Rachel |title=Getting Past Polyproline Pauses | url=http://uwo.academia.edu/PaulSzpak/Papers/827788/Fish_Bone_Chemistry_and_Ultrastructure_Implications_for_Taphonomy_and_Stable_Isotope_Analysis |journal=[[Science]] |year=2013 |volume=339 |issue=6115 |pages=38–39}}</ref> |
||
The distinct side chain/amine group interactions allow proline to aid in the formation of beta turns... |
|||
The exceptional conformational rigidity of proline affects the [[secondary structure]] of proteins near a proline residue and may account for proline's higher prevalence in the proteins of [[thermophile|thermophilic]] organisms. [[Protein secondary structure]] can be described in terms of the [[dihedral angle|dihedral angles]] [[Dihedral_angle#Dihedral angles of biological molecules|φ, ψ and ω]] of the protein backbone. The cyclic structure of proline's side chain locks the angle φ at approximately −60°. |
|||
| ⚫ | Proline acts as a structural disruptor in the middle of regular [[secondary structure]] elements such as [[alpha helix|alpha helices]] and [[beta sheet]]s; however, proline is commonly found as the first residue of an [[alpha helix]] and also in the edge strands of [[beta sheet]]s. Proline is also commonly found in [[turn (biochemistry)|turns]] (another kind of secondary structure), and aids in the formation of beta turns. This may account for the curious fact that proline is usually solvent-exposed, despite having a completely [[aliphatic]] side chain. |
||
Multiple prolines and/or [[hydroxyproline]]s in a row can create a [[polyproline helix]], the predominant [[secondary structure]] in [[collagen]]. The [[hydroxylation]] of proline by [[prolyl hydroxylase]] (or other additions of electron-withdrawing substituents such as [[fluorine]]) increases the conformational stability of [[collagen]] significantly.<ref name="SzpakJAS">{{Cite journal |last=Szpak |first=Paul |title=Fish bone chemistry and ultrastructure: implications for taphonomy and stable isotope analysis | url=http://uwo.academia.edu/PaulSzpak/Papers/827788/Fish_Bone_Chemistry_and_Ultrastructure_Implications_for_Taphonomy_and_Stable_Isotope_Analysis |journal=[[Journal of Archaeological Science (journal)|Journal of Archaeological Science]] |year=2011 |volume=38 |issue=12 |pages=3358–3372 |doi=10.1016/j.jas.2011.07.022 }}</ref> Hence, the hydroxylation of proline is a critical biochemical process for maintaining the [[connective tissue]] of higher organisms. Severe diseases such as [[scurvy]] can result from defects in this hydroxylation, e.g., mutations in the enzyme prolyl hydroxylase or lack of the necessary [[vitamin C|ascorbate (vitamin C)]] cofactor. |
Multiple prolines and/or [[hydroxyproline]]s in a row can create a [[polyproline helix]], the predominant [[secondary structure]] in [[collagen]]. The [[hydroxylation]] of proline by [[prolyl hydroxylase]] (or other additions of electron-withdrawing substituents such as [[fluorine]]) increases the conformational stability of [[collagen]] significantly.<ref name="SzpakJAS">{{Cite journal |last=Szpak |first=Paul |title=Fish bone chemistry and ultrastructure: implications for taphonomy and stable isotope analysis | url=http://uwo.academia.edu/PaulSzpak/Papers/827788/Fish_Bone_Chemistry_and_Ultrastructure_Implications_for_Taphonomy_and_Stable_Isotope_Analysis |journal=[[Journal of Archaeological Science (journal)|Journal of Archaeological Science]] |year=2011 |volume=38 |issue=12 |pages=3358–3372 |doi=10.1016/j.jas.2011.07.022 }}</ref> Hence, the hydroxylation of proline is a critical biochemical process for maintaining the [[connective tissue]] of higher organisms. Severe diseases such as [[scurvy]] can result from defects in this hydroxylation, e.g., mutations in the enzyme prolyl hydroxylase or lack of the necessary [[vitamin C|ascorbate (vitamin C)]] cofactor. |
||
Sequences of proline and [[2-Aminoisobutyric acid|2-aminoisobutyric acid]] (Aib) also form a helical turn structure.{{Citation needed|date=February 2007}} |
Sequences of proline and [[2-Aminoisobutyric acid|2-aminoisobutyric acid]] (Aib) also form a helical turn structure.{{Citation needed|date=February 2007}} |
||
| ⚫ | Peptide bond formation with incoming Pro-tRNA<sup>Pro</sup> is considerably slower than with any other tRNAs, which is a general feature of N-alkylamino acids.<ref>{{citation | first1 = Michael Y | last1 = Pavlov | first2 = Richard E | last2 = Watts | first3 = Zhongping | last3 = Tan | first4 = Virginia W | last4 = Cornish | first5 = Måns | last5 = Ehrenberg | first6 = Anthony C | title = Slow peptide bond formation by proline and other N-alkylamino acids in translation | year = 2010 | journal = PNAS | volume = 106 | issue = 1 | pages = 50–54 | doi = 10.1073/pnas.0809211106 | pmid = 19104062 | last6 = Forster | pmc = 2629218}}.</ref> Peptide bond formation is also slow between an incoming tRNA and a chain ending in |
||
==Cis-trans isomerization== |
==Cis-trans isomerization== |
||
Revision as of 00:03, 14 December 2013
| |||
| Names | |||
|---|---|---|---|
| IUPAC name
Proline
| |||
| Systematic IUPAC name
Pyrrolidine-2-carboxylic acid[1] | |||
| Identifiers | |||
3D model (JSmol)
|
|||
| 80812 | |||
| ChEBI | |||
| ChEMBL | |||
| ChemSpider | |||
| DrugBank | |||
| ECHA InfoCard | 100.009.264 | ||
| EC Number |
| ||
| 26927 | |||
| KEGG | |||
| MeSH | Proline | ||
PubChem CID
|
|||
| RTECS number |
| ||
| UNII | |||
CompTox Dashboard (EPA)
|
|||
| |||
| |||
| Properties | |||
| C5H9NO2 | |||
| Molar mass | 115.132 g·mol−1 | ||
| Appearance | Transparent crystals | ||
| Solubility | 1.5g/100g ethanol 19 degC[2] | ||
| log P | -0.06 | ||
| Acidity (pKa) | 1.99 (carboxyl), 10.96 (amino)[3] | ||
| Supplementary data page | |||
| Proline (data page) | |||
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |||
Proline (abbreviated as Pro or P) is an α-amino acid, one of the twenty DNA-encoded amino acids. Its codons are CCU, CCC, CCA, and CCG. It is not an essential amino acid, which means that the human body can synthesize it. It is unique among the 20 protein-forming amino acids in that the amine nitrogen is bound to not one but two alkyl groups, thus making it a secondary amine. The more common L form has S stereochemistry.
Biosynthesis
Proline is biosynthetically derived from the amino acid L-glutamate and its immediate precursor is the imino acid (S)-1-pyrroline-5-carboxylate (P5C). Enzymes involved in a typical biosynthesis include:[4]
- Glutamate 5-kinase, Glutamate 1-kinase (ATP-dependent)
- Glutamate dehydrogenase (requires NADH or NADPH)
- Pyrroline-5-carboxylate reductase (requires NADH or NADPH)

Properties in protein structure
The distinctive cyclic structure of proline's side chain gives proline an exceptional conformational rigidity compared to other amino acids. It also affects the rate of peptide bond formation between proline and other amino acids. When proline is bound as an amide in a peptide bond, its nitrogen is not bound to any hydrogen, meaning it cannot act as a hydrogen bond donor, but can be a hydrogen bond acceptor.
Peptide bond formation with incoming Pro-tRNAPro is considerably slower than with any other tRNAs, which is a general feature of N-alkylamino acids.[5] Peptide bond formation is also slow between an incoming tRNA and a chain ending in proline; with the creation of proline-proline bonds slowest of all.[6]
The exceptional conformational rigidity of proline affects the secondary structure of proteins near a proline residue and may account for proline's higher prevalence in the proteins of thermophilic organisms. Protein secondary structure can be described in terms of the dihedral angles φ, ψ and ω of the protein backbone. The cyclic structure of proline's side chain locks the angle φ at approximately −60°.
Proline acts as a structural disruptor in the middle of regular secondary structure elements such as alpha helices and beta sheets; however, proline is commonly found as the first residue of an alpha helix and also in the edge strands of beta sheets. Proline is also commonly found in turns (another kind of secondary structure), and aids in the formation of beta turns. This may account for the curious fact that proline is usually solvent-exposed, despite having a completely aliphatic side chain.
Multiple prolines and/or hydroxyprolines in a row can create a polyproline helix, the predominant secondary structure in collagen. The hydroxylation of proline by prolyl hydroxylase (or other additions of electron-withdrawing substituents such as fluorine) increases the conformational stability of collagen significantly.[7] Hence, the hydroxylation of proline is a critical biochemical process for maintaining the connective tissue of higher organisms. Severe diseases such as scurvy can result from defects in this hydroxylation, e.g., mutations in the enzyme prolyl hydroxylase or lack of the necessary ascorbate (vitamin C) cofactor.
Sequences of proline and 2-aminoisobutyric acid (Aib) also form a helical turn structure.[citation needed]
Cis-trans isomerization
Peptide bonds to proline, and to other N-substituted amino acids (such as sarcosine), are able to populate both the cis and trans isomers. Most peptide bonds overwhelmingly adopt the trans isomer (typically 99.9% under unstrained conditions), chiefly because the amide hydrogen (trans isomer) offers less steric repulsion to the preceding atom than does the following atom (cis isomer). By contrast, the cis and trans isomers of the X-Pro peptide bond (where X represents any amino acid) both experience steric clashes with the neighboring substitution and are nearly equal energetically. Hence, the fraction of X-Pro peptide bonds in the cis isomer under unstrained conditions ranges from 10-40%; the fraction depends slightly on the preceding amino acid, with aromatic residues favoring the cis isomer slightly.
From a kinetic standpoint, cis-trans proline isomerization is a very slow process that can impede the progress of protein folding by trapping one or more proline residues crucial for folding in the non-native isomer, especially when the native protein requires the cis isomer. This is because proline residues are exclusively synthesized in the ribosome as the trans isomer form. All organisms possess prolyl isomerase enzymes to catalyze this isomerization, and some bacteria have specialized prolyl isomerases associated with the ribosome. However, not all prolines are essential for folding, and protein folding may proceed at a normal rate despite having non-native conformers of many X-Pro peptide bonds.
Uses
Proline and its derivatives are often used as asymmetric catalysts in organic reactions. The CBS reduction and proline catalysed aldol condensation are prominent examples.
L-Proline is an osmoprotectant and therefore is used in many pharmaceutical, biotechnological applications.
In brewing, proteins rich in proline combine with polyphenols to produce haze (turbidity).[8]
Specialties
Proline is one of the two amino acids that do not follow along with the typical Ramachandran plot, along with glycine. Due to the ring formation connected to the beta carbon, the ψ and φ angles about the peptide bond have fewer allowable degrees of rotation. As a result it is often found in "turns" of proteins as its free entropy (ΔS) is not as comparatively large to other amino acids and thus in a folded form vs. unfolded form, the change in entropy is less. Furthermore, proline is rarely found in α and β structures as it would reduce the stability of such structures, because its side chain α-N can only form one hydrogen bond.
Additionally, proline is the only amino acid that does not form a blue/purple colour when developed by spraying with ninhydrin for uses in chromatography. Proline, instead, produces an orange/yellow colour.
History
Richard Willstätter synthesized proline by the reaction of sodium salt of diethyl malonate with 1,3-dibromopropane in 1900. In 1901, Hermann Emil Fischer isolated proline from casein and the decomposition products of γ-phthalimido-propylmalonic ester.[9]
See also
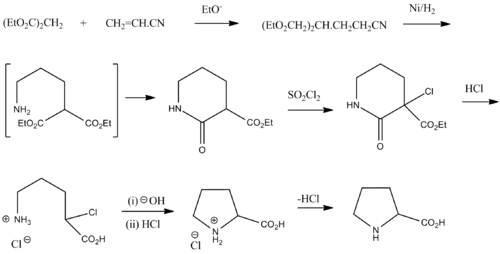
Synthesis
Racemic proline can be synthesized from diethyl malonate and acrylonitrile:[10]
References
- ^ http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=614&loc=ec_rcs
- ^ http://books.google.ch/books?id=xteiARU46SQC&pg=PA15&lpg=PA15&dq=methionine+solubility+in+ethanol&source=bl&ots=HzHueOPPoB&sig=KjMXxDNgjSvG1CddED9lfaYEhKQ&hl=en&sa=X&ei=2-26T-bZK-mX0QWt3I2ACA&redir_esc=y#v=onepage&q=methionine%20solubility%20in%20ethanol&f=false
- ^ Nelson, D.L., Cox, M.M., Principles of Biochemistry. NY: W.H. Freeman and Company.
- ^ Lehninger, Albert L.; Nelson, David L.; Cox, Michael M. (2000). Principles of Biochemistry (3rd ed.). New York: W. H. Freeman. ISBN 1-57259-153-6..
- ^ Pavlov, Michael Y; Watts, Richard E; Tan, Zhongping; Cornish, Virginia W; Ehrenberg, Måns; Forster, Anthony C (2010), "Slow peptide bond formation by proline and other N-alkylamino acids in translation", PNAS, 106 (1): 50–54, doi:10.1073/pnas.0809211106, PMC 2629218, PMID 19104062.
- ^ Buskirk, Allen R.; Green, Rachel (2013). "Getting Past Polyproline Pauses". Science. 339 (6115): 38–39.
- ^ Szpak, Paul (2011). "Fish bone chemistry and ultrastructure: implications for taphonomy and stable isotope analysis". Journal of Archaeological Science. 38 (12): 3358–3372. doi:10.1016/j.jas.2011.07.022.
- ^ K.J. Siebert, "Haze and Foam",[1] Accessed July 12, 2010.
- ^ R.H.A. Plimmer (1912) [1908], R.H.A. Plimmer & F.G. Hopkins (ed.), The chemical composition of the proteins, Monographs on biochemistry, vol. Part I. Analysis (2nd ed.), London: Longmans, Green and Co., p. 130, retrieved September 20, 2010
- ^ Vogel, Practical Organic Chemistry 5th edition
Further reading
- Balbach, J.; Schmid, F. X. (2000), "Proline isomerization and its catalysis in protein folding", in Pain, R. H. (ed.), Mechanisms of Protein Folding (2nd ed.), Oxford University Press, pp. 212–49, ISBN 0-19-963788-1.
- For a thorough scientific overview of disorders of proline and hydroxyproline metabolism, one can consult chapter 81 of OMMBID Charles Scriver, Beaudet, A.L., Valle, D., Sly, W.S., Vogelstein, B., Childs, B., Kinzler, K.W. (Accessed 2007). The Online Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill. - Summaries of 255 chapters, full text through many universities. There is also the OMMBID blog.
External links